033_OBO_LoadExtract
033_OBO_LoadExtract
Core Concepts
Compared to other numerical computing environments, Breeze matrices default to column major ordering, like Matlab, but indexing is 0-based, like Numpy. Breeze has as its core concepts matrices and column vectors. Row vectors are normally stored as matrices with a single row. This allows for greater type safety with the downside that conversion of row vectors to column vectors is performed using a transpose-slice (a.t(::,0)) instead of a simple transpose (a.t).
[[UFunc|Universal Functions]]s are very important in Breeze. Once you get a feel for the syntax (i.e. what's in this section), it might be worthwhile to read the first half of the UFunc wiki page. (You can skip the last half that involves implementing your own UFuncs...until you're ready to contribute to Breeze!)
Quick Reference
The following table assumes that Numpy is used with from numpy import * and Breeze with:
import breeze.linalg._
import breeze.numerics._
import breeze.linalg._
import breeze.numerics._
Creation
| Operation | Breeze | Matlab | Numpy | R |
|---|---|---|---|---|
| Zeroed matrix | DenseMatrix.zeros[Double](n,m) | zeros(n,m) | zeros((n,m)) | mat.or.vec(n, m) |
| Zeroed vector | DenseVector.zeros[Double](n) | zeros(n,1) | zeros(n) | mat.or.vec(n, 1) |
| Vector of ones | DenseVector.ones[Double](n) | ones(n,1) | ones(n) | mat.or.vec(n, 1) + 1 |
| Vector of particular number | DenseVector.fill(n){5.0} | ones(n,1) * 5 | ones(n) * 5 | (mat.or.vec(5, 1) + 1) * 5 |
| range given stepsize | DenseVector.range(start,stop,step) or Vector.rangeD(start,stop,step) | seq(start,stop,step) | ||
| n element range | linspace(start,stop,numvals) | linspace(0,20,15) | ||
| Identity matrix | DenseMatrix.eye[Double](n) | eye(n) | eye(n) | identity(n) |
| Diagonal matrix | diag(DenseVector(1.0,2.0,3.0)) | diag([1 2 3]) | diag((1,2,3)) | diag(c(1,2,3)) |
| Matrix inline creation | DenseMatrix((1.0,2.0), (3.0,4.0)) | [1 2; 3 4] | array([ [1,2], [3,4] ]) | matrix(c(1,2,3,4), nrow = 2, ncol = 2) |
| Column vector inline creation | DenseVector(1,2,3,4) | [1 2 3 4] | array([1,2,3,4]) | c(1,2,3,4) |
| Row vector inline creation | DenseVector(1,2,3,4).t | [1 2 3 4]' | array([1,2,3]).reshape(-1,1) | t(c(1,2,3,4)) |
| Vector from function | DenseVector.tabulate(3){i => 2*i} | |||
| Matrix from function | DenseMatrix.tabulate(3, 2){case (i, j) => i+j} | |||
| Vector creation from array | new DenseVector(Array(1, 2, 3, 4)) | |||
| Matrix creation from array | new DenseMatrix(2, 3, Array(11, 12, 13, 21, 22, 23)) | |||
| Vector of random elements from 0 to 1 | DenseVector.rand(4) | runif(4) (requires stats library) | ||
| Matrix of random elements from 0 to 1 | DenseMatrix.rand(2, 3) | matrix(runif(6),2) (requires stats library) |
DenseMatrix.zeros[Double](2,3)
res0: breeze.linalg.DenseMatrix[Double] =
0.0 0.0 0.0
0.0 0.0 0.0
import numpy as np
np.zeros((2,3))
<span class="ansired">Out[</span><span class="ansired">2</span><span class="ansired">]: </span>
array([[ 0., 0., 0.],
[ 0., 0., 0.]])
mat.or.vec(2,3)
Reading and writing Matrices
Currently, Breeze supports IO for Matrices in two ways: Java serialization and csv. The latter comes from two functions: breeze.linalg.csvread and breeze.linalg.csvwrite. csvread takes a File, and optionally parameters for how the CSV file is delimited (e.g. if it is actually a tsv file, you can set tabs as the field delimiter.) and returns a DenseMatrix. Similarly, csvwrite takes a File and a DenseMatrix, and writes the contents of a matrix to a file.
Indexing and Slicing
| Operation | Breeze | Matlab | Numpy | R |
|---|---|---|---|---|
| Basic Indexing | a(0,1) | a(1,2) | a[0,1] | a[1,2] |
| Extract subset of vector | a(1 to 4) or a(1 until 5) or a.slice(1,5) | a(2:5) | a[1:5] | a[2:5] |
| (negative steps) | a(5 to 0 by -1) | a(6:-1:1) | a[5:0:-1] | |
| (tail) | a(1 to -1) | a(2:end) | a[1:] | a[2:length(a)] or tail(a,n=length(a)-1) |
| (last element) | a( -1 ) | a(end) | a[-1] | tail(a, n=1) |
| Extract column of matrix | a(::, 2) | a(:,3) | a[:,2] | a[,2] |
val matrix = DenseMatrix.rand(2, 3)
matrix: breeze.linalg.DenseMatrix[Double] =
0.31972455666138666 0.20027601169839704 0.5602628276401904
0.4299695123127245 0.9935026349626817 0.0383067428009598
val two_one = matrix(1, 0) // Remember the index starts from zero
two_one: Double = 0.4299695123127245
Other Manipulation
| Operation | Breeze | Matlab | Numpy | R |
|---|---|---|---|---|
| Reshaping | a.reshape(3, 2) | reshape(a, 3, 2) | a.reshape(3,2) | matrix(a,nrow=3,byrow=T) |
| Flatten matrix | a.toDenseVector (Makes copy) | a(:) | a.flatten() | as.vector(a) |
| Copy lower triangle | lowerTriangular(a) | tril(a) | tril(a) | a[upper.tri(a)] <- 0 |
| Copy upper triangle | upperTriangular(a) | triu(a) | triu(a) | a[lower.tri(a)] <- 0 |
| Copy (note, no parens!!) | a.copy | np.copy(a) | ||
| Create view of matrix diagonal | diag(a) | NA | diagonal(a) (Numpy >= 1.9) | |
| Vector Assignment to subset | a(1 to 4) := 5.0 | a(2:5) = 5 | a[1:4] = 5 | a[2:5] = 5 |
| Vector Assignment to subset | a(1 to 4) := DenseVector(1.0,2.0,3.0) | a(2:5) = [1 2 3] | a[1:4] = array([1,2,3]) | a[2:5] = c(1,2,3) |
| Matrix Assignment to subset | a(1 to 3,1 to 3) := 5.0 | a(2:4,2:4) = 5 | a[1:3,1:3] = 5 | a[2:4,2:4] = 5 |
| Matrix Assignment to column | a(::, 2) := 5.0 | a(:,3) = 5 | a[:,2] = 5 | a[,3] = 5 |
| Matrix vertical concatenate | DenseMatrix.vertcat(a,b) | [a ; b] | vstack((a,b)) | rbind(a, b) |
| Matrix horizontal concatenate | DenseMatrix.horzcat(d,e) | [d , e] | hstack((d,e)) | cbind(d, e) |
| Vector concatenate | DenseVector.vertcat(a,b) | [a b] | concatenate((a,b)) | c(a, b) |
Operations
| Operation | Breeze | Matlab | Numpy | R |
|---|---|---|---|---|
| Elementwise addition | a + b | a + b | a + b | a + b |
| Shaped/Matrix multiplication | a * b | a * b | dot(a, b) | a %*% b |
| Elementwise multiplication | a :* b | a .* b | a * b | a * b |
| Elementwise division | a :/ b | a ./ b | a / b | a / b |
| Elementwise comparison | a :< b | a < b (gives matrix of 1/0 instead of true/false) | a < b | a < b |
| Elementwise equals | a :== b | a == b (gives matrix of 1/0 instead of true/false) | a == b | a == b |
| Inplace addition | a :+= 1.0 | a += 1 | a += 1 | a = a + 1 |
| Inplace elementwise multiplication | a :*= 2.0 | a *= 2 | a *= 2 | a = a * 2 |
| Vector dot product | a dot b, a.t * b† | dot(a,b) | dot(a,b) | crossprod(a,b) |
| Elementwise max | max(a) | max(a) | a.max() | max(a) |
| Elementwise argmax | argmax(a) | [v i] = max(a); i | a.argmax() | which.max(a) |
Sum
| Operation | Breeze | Matlab | Numpy | R |
|---|---|---|---|---|
| Elementwise sum | sum(a) | sum(sum(a)) | a.sum() | sum(a) |
| Sum down each column (giving a row vector) | sum(a, Axis._0) or sum(a(::, *)) | sum(a) | sum(a,0) | apply(a,2,sum) |
| Sum across each row (giving a column vector) | sum(a, Axis._1) or sum(a(*, ::)) | sum(a') | sum(a,1) | apply(a,1,sum) |
| Trace (sum of diagonal elements) | trace(a) | trace(a) | a.trace() | sum(diag(a)) |
| Cumulative sum | accumulate(a) | cumsum(a) | a.cumsum() | apply(a,2,cumsum) |
Boolean Operators
| Operation | Breeze | Matlab | Numpy | R |
|---|---|---|---|---|
| Elementwise and | a :& b | a && b | a & b | a & b |
| Elementwise or | `a : | b` | `a | |
| Elementwise not | !a | ~a | ~a | !a |
| True if any element is nonzero | any(a) | any(a) | any(a) | |
| True if all elements are nonzero | all(a) | all(a) | all(a) |
Linear Algebra Functions
| Operation | Breeze | Matlab | Numpy | R |
|---|---|---|---|---|
| Linear solve | a \ b | a \ b | linalg.solve(a,b) | solve(a,b) |
| Transpose | a.t | a' | a.conj.transpose() | t(a) |
| Determinant | det(a) | det(a) | linalg.det(a) | det(a) |
| Inverse | inv(a) | inv(a) | linalg.inv(a) | solve(a) |
| Moore-Penrose Pseudoinverse | pinv(a) | pinv(a) | linalg.pinv(a) | |
| Vector Frobenius Norm | norm(a) | norm(a) | norm(a) | |
| Eigenvalues (Symmetric) | eigSym(a) | [v,l] = eig(a) | linalg.eig(a)[0] | |
| Eigenvalues | val (er, ei, _) = eig(a) (separate real & imaginary part) | eig(a) | linalg.eig(a)[0] | eigen(a)$values |
| Eigenvectors | eig(a)._3 | [v,l] = eig(a) | linalg.eig(a)[1] | eigen(a)$vectors |
| Singular Value Decomposition | val svd.SVD(u,s,v) = svd(a) | svd(a) | linalg.svd(a) | svd(a)$d |
| Rank | rank(a) | rank(a) | rank(a) | rank(a) |
| Vector length | a.length | size(a) | a.size | length(a) |
| Matrix rows | a.rows | size(a,1) | a.shape[0] | nrow(a) |
| Matrix columns | a.cols | size(a,2) | a.shape[1] | ncol(a) |
Rounding and Signs
| Operation | Breeze | Matlab | Numpy | R |
|---|---|---|---|---|
| Round | round(a) | round(a) | around(a) | round(a) |
| Ceiling | ceil(a) | ceil(a) | ceil(a) | ceiling(a) |
| Floor | floor(a) | floor(a) | floor(a) | floor(a) |
| Sign | signum(a) | sign(a) | sign(a) | sign(a) |
| Absolute Value | abs(a) | abs(a) | abs(a) | abs(a) |
Constants
| Operation | Breeze | Matlab | Numpy | R |
|---|---|---|---|---|
| Not a Number | NaN or nan | NaN | nan | NA |
| Infinity | Inf or inf | Inf | inf | Inf |
| Pi | Constants.Pi | pi | math.pi | pi |
| e | Constants.E | exp(1) | math.e | exp(1) |
Complex numbers
If you make use of complex numbers, you will want to include a breeze.math._ import. This declares a i variable, and provides implicit conversions from Scala’s basic types to complex types.
| Operation | Breeze | Matlab | Numpy | R |
|---|---|---|---|---|
| Imaginary unit | i | i | z = 1j | 1i |
| Complex numbers | 3 + 4 * i or Complex(3,4) | 3 + 4i | z = 3 + 4j | 3 + 4i |
| Absolute Value | abs(z) or z.abs | abs(z) | abs(z) | abs(z) |
| Real Component | z.real | real(z) | z.real | Re(z) |
| Imaginary Component | z.imag | imag(z) | z.imag() | Im(z) |
| Imaginary Conjugate | z.conjugate | conj(z) | z.conj() or z.conjugate() | Conj(z) |
Numeric functions
Breeze contains a fairly comprehensive set of special functions under the breeze.numerics._ import. These functions can be applied to single elements, vectors or matrices of Doubles. This includes versions of the special functions from scala.math that can be applied to vectors and matrices. Any function acting on a basic numeric type can “vectorized”, to a [[UFunc|Universal Functions]] function, which can act elementwise on vectors and matrices:
val v = DenseVector(1.0,2.0,3.0)
exp(v) // == DenseVector(2.7182818284590455, 7.38905609893065, 20.085536923187668)
UFuncs can also be used in-place on Vectors and Matrices:
val v = DenseVector(1.0,2.0,3.0)
exp.inPlace(v) // == DenseVector(2.7182818284590455, 7.38905609893065, 20.085536923187668)
See [[Universal Functions]] for more information.
Here is a (non-exhaustive) list of UFuncs in Breeze:
Trigonometry
sin,sinh,asin,asinhcos,cosh,acos,acoshtan,tanh,atan,atanhatan2sinc(x) == sin(x)/xsincpi(x) == sinc(x * Pi)
Logarithm, Roots, and Exponentials
log,explog10log1p,expm1sqrt,sbrtpow
Gamma Function and its cousins
The gamma function is the extension of the factorial function to the reals. Numpy needs from scipy.special import * for this and subsequent sections.
| Operation | Breeze | Matlab | Numpy | R |
|---|---|---|---|---|
| Gamma function | exp(lgamma(a)) | gamma(a) | gamma(a) | gamma(a) |
| log Gamma function | lgamma(a) | gammaln(a) | gammaln(a) | lgamma(a) |
| Incomplete gamma function | gammp(a, x) | gammainc(a, x) | gammainc(a, x) | pgamma(a, x) (requires stats library) |
| Upper incomplete gamma function | gammq(a, x) | gammainc(a, x, tail) | gammaincc(a, x) | pgamma(x, a, lower = FALSE) * gamma(a) (requires stats library) |
| derivative of lgamma | digamma(a) | psi(a) | polygamma(0, a) | digamma(a) |
| derivative of digamma | trigamma(a) | psi(1, a) | polygamma(1, a) | trigama(a) |
| nth derivative of digamma | na | psi(n, a) | polygamma(n, a) | psigamma(a, deriv = n) |
| Log Beta function | lbeta(a,b) | betaln(a, b) | betaln(a,b) | lbeta(a, b) |
| Generalized Log Beta function | lbeta(a) | na | na |
Error Function
The error function...
| Operation | Breeze | Matlab | Numpy | R |
|---|---|---|---|---|
| error function | erf(a) | erf(a) | erf(a) | 2 * pnorm(a * sqrt(2)) - 1 |
| 1 - erf(a) | erfc(a) | erfc(a) | erfc(a) | 2 * pnorm(a * sqrt(2), lower = FALSE) |
| inverse error function | erfinv(a) | erfinv(a) | erfinv(a) | qnorm((1 + a) / 2) / sqrt(2) |
| inverse erfc | erfcinv(a) | erfcinv(a) | erfcinv(a) | qnorm(a / 2, lower = FALSE) / sqrt(2) |
Other functions
| Operation | Breeze | Matlab | Numpy | R |
|---|---|---|---|---|
| logistic sigmoid | sigmoid(a) | na | expit(a) | sigmoid(a) (requires pracma library) |
| Indicator function | I(a) | not needed | where(cond, 1, 0) | 0 + (a > 0) |
| Polynominal evaluation | polyval(coef,x) |
Map and Reduce
For most simple mapping tasks, one can simply use vectorized, or universal functions. Given a vector v, we can simply take the log of each element of a vector with log(v). Sometimes, however, we want to apply a somewhat idiosyncratic function to each element of a vector. For this, we can use the map function:
val v = DenseVector(1.0,2.0,3.0)
v.map( xi => foobar(xi) )
Breeze provides a number of built in reduction functions such as sum, mean. You can implement a custom reduction using the higher order function reduce. For instance, we can sum the first 9 integers as follows:
val v = linspace(0,9,10)
val s = v.reduce( _ + _ )
Broadcasting
Sometimes we want to apply an operation to every row or column of a matrix, as a unit. For instance, you might want to compute the mean of each row, or add a vector to every column. Adapting a matrix so that operations can be applied columnwise or rowwise is called broadcasting. Languages like R and numpy automatically and implicitly do broadcasting, meaning they won’t stop you if you accidentally add a matrix and a vector. In Breeze, you have to signal your intent using the broadcasting operator *. The * is meant to evoke “foreach” visually. Here are some examples:
val dm = DenseMatrix((1.0,2.0,3.0),
(4.0,5.0,6.0))
val res = dm(::, *) + DenseVector(3.0, 4.0)
assert(res === DenseMatrix((4.0, 5.0, 6.0), (8.0, 9.0, 10.0)))
res(::, *) := DenseVector(3.0, 4.0)
assert(res === DenseMatrix((3.0, 3.0, 3.0), (4.0, 4.0, 4.0)))
val m = DenseMatrix((1.0, 3.0), (4.0, 4.0))
// unbroadcasted sums all elements
assert(sum(m) === 12.0)
assert(mean(m) === 3.0)
assert(sum(m(*, ::)) === DenseVector(4.0, 8.0))
assert(sum(m(::, *)) === DenseMatrix((5.0, 7.0)))
assert(mean(m(*, ::)) === DenseVector(2.0, 4.0))
assert(mean(m(::, *)) === DenseMatrix((2.5, 3.5)))
The UFunc trait is similar to numpy’s ufunc. See [[Universal Functions]] for more information on Breeze UFuncs.
Casting and type safety
Compared to Numpy and Matlab, Breeze requires you to be more explicit about the types of your variables. When you create a new vector for example, you must specify a type (such as in DenseVector.zeros[Double](n)) in cases where a type can not be inferred automatically. Automatic inference will occur when you create a vector by passing its initial values in (DenseVector). A common mistake is using integers for initialisation (e.g. DenseVector), which would give a matrix of integers instead of doubles. Both Numpy and Matlab would default to doubles instead.
Breeze will not convert integers to doubles for you in most expressions. Simple operations like a :+ 3 when a is a DenseVector[Double] will not compile. Breeze provides a convert function, which can be used to explicitly cast. You can also use v.mapValues(_.toDouble).
Casting
| Operation | Breeze | Matlab | Numpy | R |
|---|---|---|---|---|
| Convert to Int | convert(a, Int) | int(a) | a.astype(int) | as.integer(a) |
Performance
Breeze uses netlib-java for its core linear algebra routines. This includes all the cubic time operations, matrix-matrix and matrix-vector multiplication. Special efforts are taken to ensure that arrays are not copied.
Netlib-java will attempt to load system optimised BLAS/LAPACK if they are installed, falling back to the reference natives, falling back to pure Java. Set your logger settings to ALL for the com.github.fommil.netlib package to check the status, and to com.github.fommil.jniloader for a more detailed breakdown. Read the netlib-java project page for more details.
Currently vectors and matrices over types other than Double, Float and Int are boxed, so they will typically be a lot slower. If you find yourself needing other AnyVal types like Long or Short, please ask on the list about possibly adding support for them.
This is an elaboration of the small subset of Apache Spark 2.2 mllib-progamming-guide that one needs to dive a bit deeper into distributed linear algebra.
This is a huge task to complete for the entire mlib-programming-guide. Perhaps worth continuing for Spark 2.2. Any contributions in this 'databricksification' of the programming guide are most welcome. Please feel free to send pull-requests or just fork and push yourself at https://github.com/raazesh-sainudiin/scalable-data-science.
Overview
- Data Types - MLlib Programming Guide
- Local vector and URL
- Labeled point and URL
- Local matrix and URL
- Distributed matrix and URL
- RowMatrix and URL
- IndexedRowMatrix and URL
- CoordinateMatrix and URL
- BlockMatrix and URL
This is an elaboration of the Apache Spark mllib-progamming-guide on mllib-data-types.
Overview
Data Types - MLlib Programming Guide
- Local vector and URL
- Labeled point and URL
- Local matrix and URL
- Distributed matrix and URL
- RowMatrix and URL
- IndexedRowMatrix and URL
- CoordinateMatrix and URL
- BlockMatrix and URL
MLlib supports local vectors and matrices stored on a single machine, as well as distributed matrices backed by one or more RDDs. Local vectors and local matrices are simple data models that serve as public interfaces. The underlying linear algebra operations are provided by Breeze and jblas. A training example used in supervised learning is called a “labeled point” in MLlib.
This is an elaboration of the Apache Spark mllib-progamming-guide on mllib-data-types.
Overview
Data Types - MLlib Programming Guide
- Local vector and URL
- Labeled point and URL
- Local matrix and URL
- Distributed matrix and URL
- RowMatrix and URL
- IndexedRowMatrix and URL
- CoordinateMatrix and URL
- BlockMatrix and URL
MLlib supports local vectors and matrices stored on a single machine, as well as distributed matrices backed by one or more RDDs. Local vectors and local matrices are simple data models that serve as public interfaces. The underlying linear algebra operations are provided by Breeze and jblas. A training example used in supervised learning is called a “labeled point” in MLlib.
Local vector in Scala
A local vector has integer-typed and 0-based indices and double-typed values, stored on a single machine.
MLlib supports two types of local vectors: * dense and * sparse.
A dense vector is backed by a double array representing its entry values, while a sparse vector is backed by two parallel arrays: indices and values.
For example, a vector (1.0, 0.0, 3.0) can be represented: * in dense format as [1.0, 0.0, 3.0] or * in sparse format as (3, [0, 2], [1.0, 3.0]), where 3 is the size of the vector.
The base class of local vectors is Vector, and we provide two implementations: DenseVector and SparseVector. We recommend using the factory methods implemented in Vectors to create local vectors. Refer to the Vector Scala docs and Vectors Scala docs for details on the API.
import org.apache.spark.mllib.linalg.{Vector, Vectors}
// Create a dense vector (1.0, 0.0, 3.0).
val dv: Vector = Vectors.dense(1.0, 0.0, 3.0)
import org.apache.spark.mllib.linalg.{Vector, Vectors}
dv: org.apache.spark.mllib.linalg.Vector = [1.0,0.0,3.0]
// Create a sparse vector (1.0, 0.0, 3.0) by specifying its indices and values corresponding to nonzero entries.
val sv1: Vector = Vectors.sparse(3, Array(0, 2), Array(1.0, 3.0))
sv1: org.apache.spark.mllib.linalg.Vector = (3,[0,2],[1.0,3.0])
// Create a sparse vector (1.0, 0.0, 3.0) by specifying its nonzero entries.
val sv2: Vector = Vectors.sparse(3, Seq((0, 1.0), (2, 3.0)))
sv2: org.apache.spark.mllib.linalg.Vector = (3,[0,2],[1.0,3.0])
Note: Scala imports scala.collection.immutable.Vector by default, so you have to import org.apache.spark.mllib.linalg.Vector explicitly to use MLlib’s Vector.
python: MLlib recognizes the following types as dense vectors:
- NumPy’s
array - Python’s list, e.g.,
[1, 2, 3]
and the following as sparse vectors:
- MLlib’s
SparseVector. - SciPy’s
csc_matrixwith a single column
We recommend using NumPy arrays over lists for efficiency, and using the factory methods implemented in Vectors to create sparse vectors.
Refer to the Vectors Python docs for more details on the API.
import numpy as np
import scipy.sparse as sps
from pyspark.mllib.linalg import Vectors
# Use a NumPy array as a dense vector.
dv1 = np.array([1.0, 0.0, 3.0])
# Use a Python list as a dense vector.
dv2 = [1.0, 0.0, 3.0]
# Create a SparseVector.
sv1 = Vectors.sparse(3, [0, 2], [1.0, 3.0])
# Use a single-column SciPy csc_matrix as a sparse vector.
sv2 = sps.csc_matrix((np.array([1.0, 3.0]), np.array([0, 2]), np.array([0, 2])), shape = (3, 1))
print dv1
print dv2
print sv1
print sv2
[ 1. 0. 3.]
[1.0, 0.0, 3.0]
(3,[0,2],[1.0,3.0])
(0, 0) 1.0
(2, 0) 3.0
This is an elaboration of the Apache Spark mllib-progamming-guide on mllib-data-types.
Overview
Data Types - MLlib Programming Guide
- Local vector and URL
- Labeled point and URL
- Local matrix and URL
- Distributed matrix and URL
- RowMatrix and URL
- IndexedRowMatrix and URL
- CoordinateMatrix and URL
- BlockMatrix and URL
MLlib supports local vectors and matrices stored on a single machine, as well as distributed matrices backed by one or more RDDs. Local vectors and local matrices are simple data models that serve as public interfaces. The underlying linear algebra operations are provided by Breeze and jblas. A training example used in supervised learning is called a “labeled point” in MLlib.
Labeled point in Scala
A labeled point is a local vector, either dense or sparse, associated with a label/response. In MLlib, labeled points are used in supervised learning algorithms.
We use a double to store a label, so we can use labeled points in both regression and classification.
For binary classification, a label should be either 0 (negative) or 1 (positive). For multiclass classification, labels should be class indices starting from zero: 0, 1, 2, ....
A labeled point is represented by the case class LabeledPoint.
Refer to the LabeledPoint Scala docs for details on the API.
//import first
import org.apache.spark.mllib.linalg.Vectors
import org.apache.spark.mllib.regression.LabeledPoint
import org.apache.spark.mllib.linalg.Vectors
import org.apache.spark.mllib.regression.LabeledPoint
// Create a labeled point with a "positive" label and a dense feature vector.
val pos = LabeledPoint(1.0, Vectors.dense(1.0, 0.0, 3.0))
pos: org.apache.spark.mllib.regression.LabeledPoint = (1.0,[1.0,0.0,3.0])
// Create a labeled point with a "negative" label and a sparse feature vector.
val neg = LabeledPoint(0.0, Vectors.sparse(3, Array(0, 2), Array(1.0, 3.0)))
neg: org.apache.spark.mllib.regression.LabeledPoint = (0.0,(3,[0,2],[1.0,3.0]))
Sparse data in Scala
It is very common in practice to have sparse training data. MLlib supports reading training examples stored in LIBSVM format, which is the default format used by LIBSVM and LIBLINEAR. It is a text format in which each line represents a labeled sparse feature vector using the following format:
label index1:value1 index2:value2 ...
where the indices are one-based and in ascending order. After loading, the feature indices are converted to zero-based.
MLUtils.loadLibSVMFile reads training examples stored in LIBSVM format.
Refer to the MLUtils Scala docs for details on the API.
import org.apache.spark.mllib.regression.LabeledPoint
import org.apache.spark.mllib.util.MLUtils
import org.apache.spark.rdd.RDD
//val examples: RDD[LabeledPoint] = MLUtils.loadLibSVMFile(sc, "data/mllib/sample_libsvm_data.txt") // from prog guide but no such data here - can wget from github
import org.apache.spark.mllib.regression.LabeledPoint
import org.apache.spark.mllib.util.MLUtils
import org.apache.spark.rdd.RDD
Load MNIST training and test datasets
Our datasets are vectors of pixels representing images of handwritten digits. For example:


display(dbutils.fs.ls("/databricks-datasets/mnist-digits/data-001/mnist-digits-train.txt"))
| path | name | size |
|---|---|---|
| dbfs:/databricks-datasets/mnist-digits/data-001/mnist-digits-train.txt | mnist-digits-train.txt | 6.9430283e7 |
val examples: RDD[LabeledPoint] = MLUtils.loadLibSVMFile(sc, "/databricks-datasets/mnist-digits/data-001/mnist-digits-train.txt")
examples: org.apache.spark.rdd.RDD[org.apache.spark.mllib.regression.LabeledPoint] = MapPartitionsRDD[69] at map at MLUtils.scala:84
examples.take(1)
res1: Array[org.apache.spark.mllib.regression.LabeledPoint] = Array((5.0,(780,[152,153,154,155,156,157,158,159,160,161,162,163,176,177,178,179,180,181,182,183,184,185,186,187,188,189,190,191,203,204,205,206,207,208,209,210,211,212,213,214,215,216,217,218,231,232,233,234,235,236,237,238,239,240,241,260,261,262,263,264,265,266,268,269,289,290,291,292,293,319,320,321,322,347,348,349,350,376,377,378,379,380,381,405,406,407,408,409,410,434,435,436,437,438,439,463,464,465,466,467,493,494,495,496,518,519,520,521,522,523,524,544,545,546,547,548,549,550,551,570,571,572,573,574,575,576,577,578,596,597,598,599,600,601,602,603,604,605,622,623,624,625,626,627,628,629,630,631,648,649,650,651,652,653,654,655,656,657,676,677,678,679,680,681,682,683],[3.0,18.0,18.0,18.0,126.0,136.0,175.0,26.0,166.0,255.0,247.0,127.0,30.0,36.0,94.0,154.0,170.0,253.0,253.0,253.0,253.0,253.0,225.0,172.0,253.0,242.0,195.0,64.0,49.0,238.0,253.0,253.0,253.0,253.0,253.0,253.0,253.0,253.0,251.0,93.0,82.0,82.0,56.0,39.0,18.0,219.0,253.0,253.0,253.0,253.0,253.0,198.0,182.0,247.0,241.0,80.0,156.0,107.0,253.0,253.0,205.0,11.0,43.0,154.0,14.0,1.0,154.0,253.0,90.0,139.0,253.0,190.0,2.0,11.0,190.0,253.0,70.0,35.0,241.0,225.0,160.0,108.0,1.0,81.0,240.0,253.0,253.0,119.0,25.0,45.0,186.0,253.0,253.0,150.0,27.0,16.0,93.0,252.0,253.0,187.0,249.0,253.0,249.0,64.0,46.0,130.0,183.0,253.0,253.0,207.0,2.0,39.0,148.0,229.0,253.0,253.0,253.0,250.0,182.0,24.0,114.0,221.0,253.0,253.0,253.0,253.0,201.0,78.0,23.0,66.0,213.0,253.0,253.0,253.0,253.0,198.0,81.0,2.0,18.0,171.0,219.0,253.0,253.0,253.0,253.0,195.0,80.0,9.0,55.0,172.0,226.0,253.0,253.0,253.0,253.0,244.0,133.0,11.0,136.0,253.0,253.0,253.0,212.0,135.0,132.0,16.0])))
Display our data. Each image has the true label (the label column) and a vector of features which represent pixel intensities (see below for details of what is in training).
display(examples.toDF) // covert to DataFrame and display for convenient db visualization
The pixel intensities are represented in features as a sparse vector, for example the first observation, as seen in row 1 of the output to display(training) below, has label as 5, i.e. the hand-written image is for the number 5. And this hand-written image is the following sparse vector (just click the triangle to the left of the feature in first row to see the following):
type: 0
size: 780
indices: [152,153,155,...,682,683]
values: [3, 18, 18,18,126,...,132,16]
Here * type: 0 says we hve a sparse vector. * size: 780 says the vector has 780 indices in total * these indices from 0,...,779 are a unidimensional indexing of the two-dimensional array of pixels in the image * indices: [152,153,155,...,682,683] are the indices from the [0,1,...,779] possible indices with non-zero values * a value is an integer encoding the gray-level at the pixel index * values: [3, 18, 18,18,126,...,132,16] are the actual gray level values, for example: * at pixed index 152 the gray-level value is 3, * at index 153 the gray-level value is 18, * ..., and finally at * at index 683 the gray-level value is 18
We could also use the following method as done in notebook 016_* already.
val training = spark.read.format("libsvm")
.option("numFeatures", "780")
.load("/databricks-datasets/mnist-digits/data-001/mnist-digits-train.txt")
training: org.apache.spark.sql.DataFrame = [label: double, features: vector]
display(training)
Labeled point in Python
A labeled point is represented by LabeledPoint.
Refer to the LabeledPoint Python docs for more details on the API.
# import first
from pyspark.mllib.linalg import SparseVector
from pyspark.mllib.regression import LabeledPoint
# Create a labeled point with a positive label and a dense feature vector.
pos = LabeledPoint(1.0, [1.0, 0.0, 3.0])
# Create a labeled point with a negative label and a sparse feature vector.
neg = LabeledPoint(0.0, SparseVector(3, [0, 2], [1.0, 3.0]))
Sparse data in Python
MLUtils.loadLibSVMFile reads training examples stored in LIBSVM format.
Refer to the MLUtils Python docs for more details on the API.
from pyspark.mllib.util import MLUtils
# examples = MLUtils.loadLibSVMFile(sc, "data/mllib/sample_libsvm_data.txt") #from prog guide but no such data here - can wget from github
examples = MLUtils.loadLibSVMFile(sc, "/databricks-datasets/mnist-digits/data-001/mnist-digits-train.txt")
examples.take(1)
res4: Array[org.apache.spark.mllib.regression.LabeledPoint] = Array((5.0,(780,[152,153,154,155,156,157,158,159,160,161,162,163,176,177,178,179,180,181,182,183,184,185,186,187,188,189,190,191,203,204,205,206,207,208,209,210,211,212,213,214,215,216,217,218,231,232,233,234,235,236,237,238,239,240,241,260,261,262,263,264,265,266,268,269,289,290,291,292,293,319,320,321,322,347,348,349,350,376,377,378,379,380,381,405,406,407,408,409,410,434,435,436,437,438,439,463,464,465,466,467,493,494,495,496,518,519,520,521,522,523,524,544,545,546,547,548,549,550,551,570,571,572,573,574,575,576,577,578,596,597,598,599,600,601,602,603,604,605,622,623,624,625,626,627,628,629,630,631,648,649,650,651,652,653,654,655,656,657,676,677,678,679,680,681,682,683],[3.0,18.0,18.0,18.0,126.0,136.0,175.0,26.0,166.0,255.0,247.0,127.0,30.0,36.0,94.0,154.0,170.0,253.0,253.0,253.0,253.0,253.0,225.0,172.0,253.0,242.0,195.0,64.0,49.0,238.0,253.0,253.0,253.0,253.0,253.0,253.0,253.0,253.0,251.0,93.0,82.0,82.0,56.0,39.0,18.0,219.0,253.0,253.0,253.0,253.0,253.0,198.0,182.0,247.0,241.0,80.0,156.0,107.0,253.0,253.0,205.0,11.0,43.0,154.0,14.0,1.0,154.0,253.0,90.0,139.0,253.0,190.0,2.0,11.0,190.0,253.0,70.0,35.0,241.0,225.0,160.0,108.0,1.0,81.0,240.0,253.0,253.0,119.0,25.0,45.0,186.0,253.0,253.0,150.0,27.0,16.0,93.0,252.0,253.0,187.0,249.0,253.0,249.0,64.0,46.0,130.0,183.0,253.0,253.0,207.0,2.0,39.0,148.0,229.0,253.0,253.0,253.0,250.0,182.0,24.0,114.0,221.0,253.0,253.0,253.0,253.0,201.0,78.0,23.0,66.0,213.0,253.0,253.0,253.0,253.0,198.0,81.0,2.0,18.0,171.0,219.0,253.0,253.0,253.0,253.0,195.0,80.0,9.0,55.0,172.0,226.0,253.0,253.0,253.0,253.0,244.0,133.0,11.0,136.0,253.0,253.0,253.0,212.0,135.0,132.0,16.0])))
This is an elaboration of the Apache Spark mllib-progamming-guide on mllib-data-types.
Overview
Data Types - MLlib Programming Guide
- Local vector and URL
- Labeled point and URL
- Local matrix and URL
- Distributed matrix and URL
- RowMatrix and URL
- IndexedRowMatrix and URL
- CoordinateMatrix and URL
- BlockMatrix and URL
MLlib supports local vectors and matrices stored on a single machine, as well as distributed matrices backed by one or more RDDs. Local vectors and local matrices are simple data models that serve as public interfaces. The underlying linear algebra operations are provided by Breeze and jblas. A training example used in supervised learning is called a “labeled point” in MLlib.
Local Matrix in Scala
A local matrix has integer-typed row and column indices and double-typed values, stored on a single machine. MLlib supports: * dense matrices, whose entry values are stored in a single double array in column-major order, and * sparse matrices, whose non-zero entry values are stored in the Compressed Sparse Column (CSC) format in column-major order.
For example, the following dense matrix: \[ \begin{pmatrix} 1.0 & 2.0 \\ 3.0 & 4.0 \\ 5.0 & 6.0 \end{pmatrix} \] is stored in a one-dimensional array [1.0, 3.0, 5.0, 2.0, 4.0, 6.0] with the matrix size (3, 2).
The base class of local matrices is Matrix, and we provide two implementations: DenseMatrix, and SparseMatrix. We recommend using the factory methods implemented in Matrices to create local matrices. Remember, local matrices in MLlib are stored in column-major order.
Refer to the Matrix Scala docs and Matrices Scala docs for details on the API.
Int.MaxValue // note the largest value an index can take
res0: Int = 2147483647
import org.apache.spark.mllib.linalg.{Matrix, Matrices}
// Create a dense matrix ((1.0, 2.0), (3.0, 4.0), (5.0, 6.0))
val dm: Matrix = Matrices.dense(3, 2, Array(1.0, 3.0, 5.0, 2.0, 4.0, 6.0))
import org.apache.spark.mllib.linalg.{Matrix, Matrices}
dm: org.apache.spark.mllib.linalg.Matrix =
1.0 2.0
3.0 4.0
5.0 6.0
Next, let us create the following sparse local matrix: \[ \begin{pmatrix} 9.0 & 0.0 \\ 0.0 & 8.0 \\ 0.0 & 6.0 \end{pmatrix} \]
// Create a sparse matrix ((9.0, 0.0), (0.0, 8.0), (0.0, 6.0))
val sm: Matrix = Matrices.sparse(3, 2, Array(0, 1, 3), Array(0, 2, 1), Array(9, 6, 8))
sm: org.apache.spark.mllib.linalg.Matrix =
3 x 2 CSCMatrix
(0,0) 9.0
(2,1) 6.0
(1,1) 8.0
Local Matrix in Python
The base class of local matrices is Matrix, and we provide two implementations: DenseMatrix, and SparseMatrix. We recommend using the factory methods implemented in Matrices to create local matrices. Remember, local matrices in MLlib are stored in column-major order.
Refer to the Matrix Python docs and Matrices Python docs for more details on the API.
from pyspark.mllib.linalg import Matrix, Matrices
# Create a dense matrix ((1.0, 2.0), (3.0, 4.0), (5.0, 6.0))
dm2 = Matrices.dense(3, 2, [1, 2, 3, 4, 5, 6])
dm2
<span class="ansired">Out[</span><span class="ansired">1</span><span class="ansired">]: </span>DenseMatrix(3, 2, [1.0, 2.0, 3.0, 4.0, 5.0, 6.0], False)
# Create a sparse matrix ((9.0, 0.0), (0.0, 8.0), (0.0, 6.0))
sm = Matrices.sparse(3, 2, [0, 1, 3], [0, 2, 1], [9, 6, 8])
sm
<span class="ansired">Out[</span><span class="ansired">2</span><span class="ansired">]: </span>SparseMatrix(3, 2, [0, 1, 3], [0, 2, 1], [9.0, 6.0, 8.0], False)
This is an elaboration of the Apache Spark mllib-progamming-guide on mllib-data-types.
Overview
Data Types - MLlib Programming Guide
- Local vector and URL
- Labeled point and URL
- Local matrix and URL
- Distributed matrix and URL
- RowMatrix and URL
- IndexedRowMatrix and URL
- CoordinateMatrix and URL
- BlockMatrix and URL
MLlib supports local vectors and matrices stored on a single machine, as well as distributed matrices backed by one or more RDDs. Local vectors and local matrices are simple data models that serve as public interfaces. The underlying linear algebra operations are provided by Breeze and jblas. A training example used in supervised learning is called a “labeled point” in MLlib.
Distributed matrix in Scala
A distributed matrix has long-typed row and column indices and double-typed values, stored distributively in one or more RDDs.
It is very important to choose the right format to store large and distributed matrices. Converting a distributed matrix to a different format may require a global shuffle, which is quite expensive.
Three types of distributed matrices have been implemented so far.
- The basic type is called
RowMatrix.
- A
RowMatrixis a row-oriented distributed matrix without meaningful row indices, e.g., a collection of feature vectors. It is backed by an RDD of its rows, where each row is a local vector. - We assume that the number of columns is not huge for a
RowMatrixso that a single local vector can be reasonably communicated to the driver and can also be stored / operated on using a single node. - An
IndexedRowMatrixis similar to aRowMatrixbut with row indices, which can be used for identifying rows and executing joins. - A
CoordinateMatrixis a distributed matrix stored in coordinate list (COO) format, backed by an RDD of its entries.
Note
The underlying RDDs of a distributed matrix must be deterministic, because we cache the matrix size. In general the use of non-deterministic RDDs can lead to errors.
Remark: there is a huge difference in the orders of magnitude between the maximum size of local versus distributed matrices!
print(Long.MaxValue.toDouble, Int.MaxValue.toDouble, Long.MaxValue.toDouble / Int.MaxValue.toDouble) // index ranges and ratio for local and distributed matrices
This is an elaboration of the Apache Spark mllib-progamming-guide on mllib-data-types.
Overview
Data Types - MLlib Programming Guide
- Local vector and URL
- Labeled point and URL
- Local matrix and URL
- Distributed matrix and URL
- RowMatrix and URL
- IndexedRowMatrix and URL
- CoordinateMatrix and URL
- BlockMatrix and URL
MLlib supports local vectors and matrices stored on a single machine, as well as distributed matrices backed by one or more RDDs. Local vectors and local matrices are simple data models that serve as public interfaces. The underlying linear algebra operations are provided by Breeze and jblas. A training example used in supervised learning is called a “labeled point” in MLlib.
RowMatrix in Scala
A RowMatrix is a row-oriented distributed matrix without meaningful row indices, backed by an RDD of its rows, where each row is a local vector. Since each row is represented by a local vector, the number of columns is limited by the integer range but it should be much smaller in practice.
A RowMatrix can be created from an RDD[Vector] instance. Then we can compute its column summary statistics and decompositions.
- QR decomposition is of the form A = QR where Q is an orthogonal matrix and R is an upper triangular matrix.
- For singular value decomposition (SVD) and principal component analysis (PCA), please refer to Dimensionality reduction.
Refer to the RowMatrix Scala docs for details on the API.
import org.apache.spark.mllib.linalg.{Vector, Vectors}
import org.apache.spark.mllib.linalg.distributed.RowMatrix
import org.apache.spark.mllib.linalg.{Vector, Vectors}
import org.apache.spark.mllib.linalg.distributed.RowMatrix
val rows: RDD[Vector] = sc.parallelize(Array(Vectors.dense(12.0, -51.0, 4.0), Vectors.dense(6.0, 167.0, -68.0), Vectors.dense(-4.0, 24.0, -41.0))) // an RDD of local vectors
rows: org.apache.spark.rdd.RDD[org.apache.spark.mllib.linalg.Vector] = ParallelCollectionRDD[18] at parallelize at <console>:36
// Create a RowMatrix from an RDD[Vector].
val mat: RowMatrix = new RowMatrix(rows)
mat: org.apache.spark.mllib.linalg.distributed.RowMatrix = org.apache.spark.mllib.linalg.distributed.RowMatrix@720029a1
mat.rows.collect
res0: Array[org.apache.spark.mllib.linalg.Vector] = Array([12.0,-51.0,4.0], [6.0,167.0,-68.0], [-4.0,24.0,-41.0])
// Get its size.
val m = mat.numRows()
val n = mat.numCols()
m: Long = 3
n: Long = 3
// QR decomposition
val qrResult = mat.tallSkinnyQR(true)
qrResult: org.apache.spark.mllib.linalg.QRDecomposition[org.apache.spark.mllib.linalg.distributed.RowMatrix,org.apache.spark.mllib.linalg.Matrix] =
QRDecomposition(org.apache.spark.mllib.linalg.distributed.RowMatrix@299d426,14.0 21.0 -14.0
0.0 -174.99999999999997 70.00000000000001
0.0 0.0 -35.000000000000014 )
qrResult.R
res1: org.apache.spark.mllib.linalg.Matrix =
14.0 21.0 -14.0
0.0 -174.99999999999997 70.00000000000001
0.0 0.0 -35.000000000000014
RowMatrix in Python
A RowMatrix can be created from an RDD of vectors.
Refer to the RowMatrix Python docs for more details on the API.
from pyspark.mllib.linalg.distributed import RowMatrix
# Create an RDD of vectors.
rows = sc.parallelize([[1, 2, 3], [4, 5, 6], [7, 8, 9], [10, 11, 12]])
# Create a RowMatrix from an RDD of vectors.
mat = RowMatrix(rows)
# Get its size.
m = mat.numRows() # 4
n = mat.numCols() # 3
print m,'x',n
# Get the rows as an RDD of vectors again.
rowsRDD = mat.rows
4 x 3
This is an elaboration of the Apache Spark mllib-progamming-guide on mllib-data-types.
Overview
Data Types - MLlib Programming Guide
- Local vector and URL
- Labeled point and URL
- Local matrix and URL
- Distributed matrix and URL
- RowMatrix and URL
- IndexedRowMatrix and URL
- CoordinateMatrix and URL
- BlockMatrix and URL
MLlib supports local vectors and matrices stored on a single machine, as well as distributed matrices backed by one or more RDDs. Local vectors and local matrices are simple data models that serve as public interfaces. The underlying linear algebra operations are provided by Breeze and jblas. A training example used in supervised learning is called a “labeled point” in MLlib.
IndexedRowMatrix in Scala
An IndexedRowMatrix is similar to a RowMatrix but with meaningful row indices. It is backed by an RDD of indexed rows, so that each row is represented by its index (long-typed) and a local vector.
An IndexedRowMatrix can be created from an RDD[IndexedRow] instance, where IndexedRow is a wrapper over (Long, Vector). An IndexedRowMatrix can be converted to a RowMatrix by dropping its row indices.
Refer to the IndexedRowMatrix Scala docs for details on the API.
import org.apache.spark.mllib.linalg.{Vector, Vectors}
import org.apache.spark.mllib.linalg.distributed.{IndexedRow, IndexedRowMatrix, RowMatrix}
import org.apache.spark.mllib.linalg.{Vector, Vectors}
import org.apache.spark.mllib.linalg.distributed.{IndexedRow, IndexedRowMatrix, RowMatrix}
Vector(12.0, -51.0, 4.0) // note Vector is a scala collection
res8: scala.collection.immutable.Vector[Double] = Vector(12.0, -51.0, 4.0)
Vectors.dense(12.0, -51.0, 4.0) // while this is a mllib.linalg.Vector
res9: org.apache.spark.mllib.linalg.Vector = [12.0,-51.0,4.0]
val rows: RDD[IndexedRow] = sc.parallelize(Array(IndexedRow(2, Vectors.dense(1,3)), IndexedRow(4, Vectors.dense(4,5)))) // an RDD of indexed rows
rows: org.apache.spark.rdd.RDD[org.apache.spark.mllib.linalg.distributed.IndexedRow] = ParallelCollectionRDD[252] at parallelize at <console>:41
// Create an IndexedRowMatrix from an RDD[IndexedRow].
val mat: IndexedRowMatrix = new IndexedRowMatrix(rows)
mat: org.apache.spark.mllib.linalg.distributed.IndexedRowMatrix = org.apache.spark.mllib.linalg.distributed.IndexedRowMatrix@2a57e8ca
// Get its size.
val m = mat.numRows()
val n = mat.numCols()
m: Long = 5
n: Long = 2
// Drop its row indices.
val rowMat: RowMatrix = mat.toRowMatrix()
rowMat: org.apache.spark.mllib.linalg.distributed.RowMatrix = org.apache.spark.mllib.linalg.distributed.RowMatrix@37fba875
rowMat.rows.collect()
res11: Array[org.apache.spark.mllib.linalg.Vector] = Array([1.0,3.0], [4.0,5.0])
IndexedRowMatrix in Python
An IndexedRowMatrix can be created from an RDD of IndexedRows, where IndexedRow is a wrapper over (long, vector). An IndexedRowMatrix can be converted to a RowMatrix by dropping its row indices.
Refer to the IndexedRowMatrix Python docs for more details on the API.
from pyspark.mllib.linalg.distributed import IndexedRow, IndexedRowMatrix
# Create an RDD of indexed rows.
# - This can be done explicitly with the IndexedRow class:
indexedRows = sc.parallelize([IndexedRow(0, [1, 2, 3]),
IndexedRow(1, [4, 5, 6]),
IndexedRow(2, [7, 8, 9]),
IndexedRow(3, [10, 11, 12])])
# - or by using (long, vector) tuples:
indexedRows = sc.parallelize([(0, [1, 2, 3]), (1, [4, 5, 6]),
(2, [7, 8, 9]), (3, [10, 11, 12])])
# Create an IndexedRowMatrix from an RDD of IndexedRows.
mat = IndexedRowMatrix(indexedRows)
# Get its size.
m = mat.numRows() # 4
n = mat.numCols() # 3
print (m,n)
# Get the rows as an RDD of IndexedRows.
rowsRDD = mat.rows
# Convert to a RowMatrix by dropping the row indices.
rowMat = mat.toRowMatrix()
# Convert to a CoordinateMatrix.
coordinateMat = mat.toCoordinateMatrix()
# Convert to a BlockMatrix.
blockMat = mat.toBlockMatrix()
(4L, 3L)
This is an elaboration of the Apache Spark mllib-progamming-guide on mllib-data-types.
Overview
Data Types - MLlib Programming Guide
- Local vector and URL
- Labeled point and URL
- Local matrix and URL
- Distributed matrix and URL
- RowMatrix and URL
- IndexedRowMatrix and URL
- CoordinateMatrix and URL
- BlockMatrix and URL
MLlib supports local vectors and matrices stored on a single machine, as well as distributed matrices backed by one or more RDDs. Local vectors and local matrices are simple data models that serve as public interfaces. The underlying linear algebra operations are provided by Breeze and jblas. A training example used in supervised learning is called a “labeled point” in MLlib.
CoordinateMatrix in Scala
A CoordinateMatrix is a distributed matrix backed by an RDD of its entries. Each entry is a tuple of (i: Long, j: Long, value: Double), where i is the row index, j is the column index, and value is the entry value. A CoordinateMatrix should be used only when both dimensions of the matrix are huge and the matrix is very sparse.
A CoordinateMatrix can be created from an RDD[MatrixEntry] instance, where MatrixEntry is a wrapper over (Long, Long, Double). A CoordinateMatrix can be converted to an IndexedRowMatrix with sparse rows by calling toIndexedRowMatrix. Other computations for CoordinateMatrix are not currently supported.
Refer to the CoordinateMatrix Scala docs for details on the API.
import org.apache.spark.mllib.linalg.distributed.{CoordinateMatrix, MatrixEntry}
import org.apache.spark.mllib.linalg.distributed.{CoordinateMatrix, MatrixEntry}
val entries: RDD[MatrixEntry] = sc.parallelize(Array(MatrixEntry(0, 0, 1.2), MatrixEntry(1, 0, 2.1), MatrixEntry(6, 1, 3.7))) // an RDD of matrix entries
entries: org.apache.spark.rdd.RDD[org.apache.spark.mllib.linalg.distributed.MatrixEntry] = ParallelCollectionRDD[454] at parallelize at <console>:35
// Create a CoordinateMatrix from an RDD[MatrixEntry].
val mat: CoordinateMatrix = new CoordinateMatrix(entries)
mat: org.apache.spark.mllib.linalg.distributed.CoordinateMatrix = org.apache.spark.mllib.linalg.distributed.CoordinateMatrix@73dc93f3
// Get its size.
val m = mat.numRows()
val n = mat.numCols()
m: Long = 7
n: Long = 2
// Convert it to an IndexRowMatrix whose rows are sparse vectors.
val indexedRowMatrix = mat.toIndexedRowMatrix()
indexedRowMatrix: org.apache.spark.mllib.linalg.distributed.IndexedRowMatrix = org.apache.spark.mllib.linalg.distributed.IndexedRowMatrix@4a8e753a
indexedRowMatrix.rows.collect()
res3: Array[org.apache.spark.mllib.linalg.distributed.IndexedRow] = Array(IndexedRow(0,(2,[0],[1.2])), IndexedRow(6,(2,[1],[3.7])), IndexedRow(1,(2,[0],[2.1])))
CoordinateMatrix in Scala
A CoordinateMatrix can be created from an RDD of MatrixEntry entries, where MatrixEntry is a wrapper over (long, long, float). A CoordinateMatrix can be converted to a RowMatrix by calling toRowMatrix, or to an IndexedRowMatrix with sparse rows by calling toIndexedRowMatrix.
Refer to the CoordinateMatrix Python docs for more details on the API.
from pyspark.mllib.linalg.distributed import CoordinateMatrix, MatrixEntry
# Create an RDD of coordinate entries.
# - This can be done explicitly with the MatrixEntry class:
entries = sc.parallelize([MatrixEntry(0, 0, 1.2), MatrixEntry(1, 0, 2.1), MatrixEntry(6, 1, 3.7)])
# - or using (long, long, float) tuples:
entries = sc.parallelize([(0, 0, 1.2), (1, 0, 2.1), (2, 1, 3.7)])
# Create an CoordinateMatrix from an RDD of MatrixEntries.
mat = CoordinateMatrix(entries)
# Get its size.
m = mat.numRows() # 3
n = mat.numCols() # 2
print (m,n)
# Get the entries as an RDD of MatrixEntries.
entriesRDD = mat.entries
# Convert to a RowMatrix.
rowMat = mat.toRowMatrix()
# Convert to an IndexedRowMatrix.
indexedRowMat = mat.toIndexedRowMatrix()
# Convert to a BlockMatrix.
blockMat = mat.toBlockMatrix()
(3L, 2L)
This is an elaboration of the Apache Spark mllib-progamming-guide on mllib-data-types.
Overview
Data Types - MLlib Programming Guide
- Local vector and URL
- Labeled point and URL
- Local matrix and URL
- Distributed matrix and URL
- RowMatrix and URL
- IndexedRowMatrix and URL
- CoordinateMatrix and URL
- BlockMatrix and URL
MLlib supports local vectors and matrices stored on a single machine, as well as distributed matrices backed by one or more RDDs. Local vectors and local matrices are simple data models that serve as public interfaces. The underlying linear algebra operations are provided by Breeze and jblas. A training example used in supervised learning is called a “labeled point” in MLlib.
BlockMatrix in Scala
A BlockMatrix is a distributed matrix backed by an RDD of MatrixBlocks, where a MatrixBlock is a tuple of ((Int, Int), Matrix), where the (Int, Int) is the index of the block, and Matrix is the sub-matrix at the given index with size rowsPerBlock x colsPerBlock. BlockMatrix supports methods such as add and multiply with another BlockMatrix. BlockMatrix also has a helper function validate which can be used to check whether the BlockMatrix is set up properly.
A BlockMatrix can be most easily created from an IndexedRowMatrix or CoordinateMatrix by calling toBlockMatrix. toBlockMatrix creates blocks of size 1024 x 1024 by default. Users may change the block size by supplying the values through toBlockMatrix(rowsPerBlock, colsPerBlock).
Refer to the BlockMatrix Scala docs for details on the API.
//import org.apache.spark.mllib.linalg.{Matrix, Matrices}
import org.apache.spark.mllib.linalg.distributed.{BlockMatrix, CoordinateMatrix, MatrixEntry}
import org.apache.spark.mllib.linalg.distributed.{BlockMatrix, CoordinateMatrix, MatrixEntry}
val entries: RDD[MatrixEntry] = sc.parallelize(Array(MatrixEntry(0, 0, 1.2), MatrixEntry(1, 0, 2.1), MatrixEntry(6, 1, 3.7))) // an RDD of matrix entries
entries: org.apache.spark.rdd.RDD[org.apache.spark.mllib.linalg.distributed.MatrixEntry] = ParallelCollectionRDD[692] at parallelize at <console>:35
// Create a CoordinateMatrix from an RDD[MatrixEntry].
val coordMat: CoordinateMatrix = new CoordinateMatrix(entries)
coordMat: org.apache.spark.mllib.linalg.distributed.CoordinateMatrix = org.apache.spark.mllib.linalg.distributed.CoordinateMatrix@68f1d303
// Transform the CoordinateMatrix to a BlockMatrix
val matA: BlockMatrix = coordMat.toBlockMatrix().cache()
matA: org.apache.spark.mllib.linalg.distributed.BlockMatrix = org.apache.spark.mllib.linalg.distributed.BlockMatrix@1de2a311
// Validate whether the BlockMatrix is set up properly. Throws an Exception when it is not valid.
// Nothing happens if it is valid.
matA.validate()
// Calculate A^T A.
val ata = matA.transpose.multiply(matA)
ata: org.apache.spark.mllib.linalg.distributed.BlockMatrix = org.apache.spark.mllib.linalg.distributed.BlockMatrix@16a80e13
ata.blocks.collect()
res1: Array[((Int, Int), org.apache.spark.mllib.linalg.Matrix)] =
Array(((0,0),5.85 0.0
0.0 13.690000000000001 ))
ata.toLocalMatrix()
res3: org.apache.spark.mllib.linalg.Matrix =
5.85 0.0
0.0 13.690000000000001
BlockMatrix in Scala
A BlockMatrix can be created from an RDD of sub-matrix blocks, where a sub-matrix block is a ((blockRowIndex, blockColIndex), sub-matrix) tuple.
Refer to the BlockMatrix Python docs for more details on the API.
from pyspark.mllib.linalg import Matrices
from pyspark.mllib.linalg.distributed import BlockMatrix
# Create an RDD of sub-matrix blocks.
blocks = sc.parallelize([((0, 0), Matrices.dense(3, 2, [1, 2, 3, 4, 5, 6])),
((1, 0), Matrices.dense(3, 2, [7, 8, 9, 10, 11, 12]))])
# Create a BlockMatrix from an RDD of sub-matrix blocks.
mat = BlockMatrix(blocks, 3, 2)
# Get its size.
m = mat.numRows() # 6
n = mat.numCols() # 2
print (m,n)
# Get the blocks as an RDD of sub-matrix blocks.
blocksRDD = mat.blocks
# Convert to a LocalMatrix.
localMat = mat.toLocalMatrix()
# Convert to an IndexedRowMatrix.
indexedRowMat = mat.toIndexedRowMatrix()
# Convert to a CoordinateMatrix.
coordinateMat = mat.toCoordinateMatrix()
(6L, 2L)
This is an elaboration of the http://spark.apache.org/docs/latest/sql-programming-guide.html by Ivan Sadikov and Raazesh Sainudiin.
Any contributions in this 'databricksification' of the programming guide are most welcome. Please feel free to send pull-requests or just fork and push yourself at/from https://github.com/lamastex/scalable-data-science.
NOTE: The links that do not have standard URLs for hyper-text transfer protocol, qualified here by (http) or (https), are in general internal links and will/should work if you follow the instructions in the lectures (from the YouTube play list, watched sequential in chronological order that is linked from https://lamastex.github.io/scalable-data-science/sds/2/2/) on how to download the .dbc archive for the course and upload it into your community edition with the correctly named expected directory structures.
Spark Sql Programming Guide
- Overview
- SQL
- DataFrames
- Datasets
- Getting Started
- Starting Point: SQLContext
- Creating DataFrames
- DataFrame Operations
- Running SQL Queries Programmatically
- Creating Datasets
- Interoperating with RDDs
- Inferring the Schema Using Reflection
- Programmatically Specifying the Schema
- Data Sources
- Generic Load/Save Functions
- Manually Specifying Options
- Run SQL on files directly
- Save Modes
- Saving to Persistent Tables
- Parquet Files
- Loading Data Programmatically
- Partition Discovery
- Schema Merging
- Hive metastore Parquet table conversion
- Hive/Parquet Schema Reconciliation
- Metadata Refreshing
- Configuration
- JSON Datasets
- Hive Tables
- Interacting with Different Versions of Hive Metastore
- JDBC To Other Databases
- Troubleshooting
- Generic Load/Save Functions
- Performance Tuning
- Caching Data In Memory
- Other Configuration Options
- Distributed SQL Engine
- Running the Thrift JDBC/ODBC server
- Running the Spark SQL CLI
This is an elaboration of the http://spark.apache.org/docs/latest/sql-programming-guide.html by Ivan Sadikov and Raazesh Sainudiin.
Any contributions in this 'databricksification' of the programming guide are most welcome. Please feel free to send pull-requests or just fork and push yourself at/from https://github.com/lamastex/scalable-data-science.
NOTE: The links that do not have standard URLs for hyper-text transfer protocol, qualified here by (http) or (https), are in general internal links and will/should work if you follow the instructions in the lectures (from the YouTube play list, watched sequential in chronological order that is linked from https://lamastex.github.io/scalable-data-science/sds/2/2/) on how to download the .dbc archive for the course and upload it into your community edition with the correctly named expected directory structures.
Overview
Spark Sql Programming Guide
TODO fix internal/external links below
- Overview
- SQL
- DataFrames
- Datasets
- Getting Started
- Starting Point: SQLContext
- Creating DataFrames
- DataFrame Operations
- Running SQL Queries Programmatically
- Creating Datasets
- Interoperating with RDDs
- Inferring the Schema Using Reflection
- Programmatically Specifying the Schema
- Data Sources
- Generic Load/Save Functions
- Manually Specifying Options
- Run SQL on files directly
- Save Modes
- Saving to Persistent Tables
- Parquet Files
- Loading Data Programmatically
- Partition Discovery
- Schema Merging
- Hive metastore Parquet table conversion
- Hive/Parquet Schema Reconciliation
- Metadata Refreshing
- Configuration
- JSON Datasets
- Hive Tables
- Interacting with Different Versions of Hive Metastore
- JDBC To Other Databases
- Troubleshooting
- Generic Load/Save Functions
- Performance Tuning
- Caching Data In Memory
- Other Configuration Options
- Distributed SQL Engine
- Running the Thrift JDBC/ODBC server
- Running the Spark SQL CLI
Overview
Spark SQL is a Spark module for structured data processing. Unlike the basic Spark RDD API, the interfaces provided by Spark SQL provide Spark with more information about the structure of both the data and the computation being performed. Internally, Spark SQL uses this extra information to perform extra optimizations. There are several ways to interact with Spark SQL including:
- SQL (SQL 2003 standard compliant)
- the DataFrames API (since Spark 1.4, was generalized in Spark 2.0 and is alias for
Dataset[Row]) - the Datasets API (offers strongly-typed interface)
When computing a result the same execution engine is used, independent of which API/language you are using to express the computation. This unification means that developers can easily switch back and forth between the various APIs based on which provides the most natural way to express a given transformation.
All of the examples on this page use sample data included in the Spark distribution and can be run in the spark-shell, pyspark shell, or sparkR shell.
SQL
One use of Spark SQL is to execute SQL queries written using either a basic SQL syntax or HiveQL. Spark SQL can also be used to read data from an existing Hive installation. For more on how to configure this feature, please refer to the Hive Tables section. When running SQL from within another programming language the results will be returned as a DataFrame. You can also interact with the SQL interface using the command-line or over JDBC/ODBC.
Datasets and DataFrames
A Dataset is a distributed collection of data. Dataset is a new interface added in Spark 1.6 that provides the benefits of RDDs (strong typing, ability to use powerful lambda functions) with the benefits of Spark SQL’s optimized execution engine, which has been improved in 2.x versions. A Dataset can be constructed (http) from JVM objects and then manipulated using functional transformations (map, flatMap, filter, etc.). The Dataset API is available in Scala (http) and Java (http). Python does not have the support for the Dataset API. But due to Python’s dynamic nature, many of the benefits of the Dataset API are already available (i.e. you can access the field of a row by name naturally row.columnName). The case for R is similar.
A DataFrame is a Dataset organized into named columns. It is conceptually equivalent to a table in a relational database or a data frame in R/Python, but with richer optimizations under the hood. DataFrames can be constructed from a wide array of sources such as: structured data files, tables in Hive, external databases, or existing RDDs. The DataFrame API is available in Scala, Java, Python, and R. In Scala and Java, a DataFrame is represented by a Dataset of Rows. In the Scala API, DataFrame is simply a type alias of Dataset[Row]. While, in Java API, users need to use Dataset<Row> to represent a DataFrame.
This is an elaboration of the Apache Spark 2.2 sql-progamming-guide.
Getting Started
Spark Sql Programming Guide
- Overview
- SQL
- DataFrames
- Datasets
- Getting Started
- Starting Point: SQLContext
- Creating DataFrames
- DataFrame Operations
- Running SQL Queries Programmatically
- Creating Datasets
- Interoperating with RDDs
- Inferring the Schema Using Reflection
- Programmatically Specifying the Schema
- Data Sources
- Generic Load/Save Functions
- Manually Specifying Options
- Run SQL on files directly
- Save Modes
- Saving to Persistent Tables
- Parquet Files
- Loading Data Programmatically
- Partition Discovery
- Schema Merging
- Hive metastore Parquet table conversion
- Hive/Parquet Schema Reconciliation
- Metadata Refreshing
- Configuration
- JSON Datasets
- Hive Tables
- Interacting with Different Versions of Hive Metastore
- JDBC To Other Databases
- Troubleshooting
- Generic Load/Save Functions
- Performance Tuning
- Caching Data In Memory
- Other Configuration Options
- Distributed SQL Engine
- Running the Thrift JDBC/ODBC server
- Running the Spark SQL CLI
Getting Started
Starting Point: SQLContext
The entry point into all functionality in Spark SQL is the SparkSession class and/or SQLContext/HiveContext. Spark session is created for you as spark when you start spark-shell or pyspark. You will need to create SparkSession usually when building an application (running on production-like on-premises cluster). n this case follow code below to create Spark session.
import org.apache.spark.sql.SparkSession
val spark = SparkSession.builder().appName("Spark SQL basic example").getOrCreate()
// you could get SparkContext and SQLContext from SparkSession
val sc = spark.sparkContext
val sqlContext = spark.sqlContext
// This is used to implicitly convert an RDD or Seq to a DataFrame (see examples below)
import spark.implicits._
But in Databricks notebook (similar to spark-shell) SparkSession is already created for you and is available as spark.
// Evaluation of the cell by Ctrl+Enter will print spark session available in notebook
spark
res0: org.apache.spark.sql.SparkSession = org.apache.spark.sql.SparkSession@2d0c6c9
After evaluation you should see something like this:
res0: org.apache.spark.sql.SparkSession = org.apache.spark.sql.SparkSession@2d0c6c9
In order to enable Hive support use enableHiveSupport() method on builder when constructing Spark session, which provides richer functionality over standard Spark SQL context, for example, usage of Hive user-defined functions or loading and writing data from/into Hive. Note that most of the SQL functionality is available regardless Hive support.
Creating DataFrames
With a SparkSessions, applications can create Dataset or DataFrame from an existing RDD, from a Hive table, or from various datasources.
Just to recap, a DataFrame is a distributed collection of data organized into named columns. You can think of it as an organized into table RDD of case class Row (which is not exactly true). DataFrames, in comparison to RDDs, are backed by rich optimizations, including tracking their own schema, adaptive query execution, code generation including whole stage codegen, extensible Catalyst optimizer, and project Tungsten.
Dataset provides type-safety when working with SQL, since Row is mapped to a case class, so that each column can be referenced by property of that class.
Note that performance for Dataset/DataFrames is the same across languages Scala, Java, Python, and R. This is due to the fact that the planning phase is just language-specific, only logical plan is constructed in Python, and all the physical execution is compiled and executed as JVM bytecode.
// Spark has some of the pre-built methods to create simple Dataset/DataFrame
// 1. Empty Dataset/DataFrame, not really interesting, is it?
println(spark.emptyDataFrame)
println(spark.emptyDataset[Int])
[]
[value: int]
// 2. Range of numbers, note that Spark automatically names column as "id"
val range = spark.range(0, 10)
// In order to get a preview of data in DataFrame use "show()"
range.show(3)
+---+
| id|
+---+
| 0|
| 1|
| 2|
+---+
only showing top 3 rows
range: org.apache.spark.sql.Dataset[Long] = [id: bigint]
You can also use different datasources that will be shown later or load Hive tables directly into Spark.
We have already created a table of social media usage from NYC (you will see later how this table was built from raw data).
See the very bottom of this worksheet to see how this was done.
First let's make sure this table is available for us.
// Let's find out what tables are already available for loading
spark.catalog.listTables.show()
+--------------------+--------+-----------+---------+-----------+
| name|database|description|tableType|isTemporary|
+--------------------+--------+-----------+---------+-----------+
| cities_csv| default| null| EXTERNAL| false|
| cleaned_taxes| default| null| MANAGED| false|
|commdettrumpclint...| default| null| MANAGED| false|
| donaldtrumptweets| default| null| EXTERNAL| false|
| linkage| default| null| EXTERNAL| false|
| nations| default| null| EXTERNAL| false|
| newmplist| default| null| EXTERNAL| false|
| ny_baby_names| default| null| MANAGED| false|
| nzmpsandparty| default| null| EXTERNAL| false|
| pos_neg_category| default| null| EXTERNAL| false|
| rna| default| null| MANAGED| false|
| samh| default| null| EXTERNAL| false|
| social_media_usage| default| null| EXTERNAL| false|
| table1| default| null| EXTERNAL| false|
| test_table| default| null| EXTERNAL| false|
| uscites| default| null| EXTERNAL| false|
+--------------------+--------+-----------+---------+-----------+
It looks like the table social_media_usage is available as a permanent table (isTemporary set as false).
Next let us do the following: * load this table as a DataFrame * print its schema and * show the first 20 rows.
val df = spark.table("social_media_usage") // Ctrl+Enter
df: org.apache.spark.sql.DataFrame = [agency: string, platform: string ... 3 more fields]
As you can see the immutable value df is a DataFrame and more specifically it is:
org.apache.spark.sql.DataFrame = [agency: string, platform: string, url: string, visits: int].
Now let us print schema of the DataFrame df and have a look at the actual data:
// Ctrl+Enter
df.printSchema() // prints schema of the DataFrame
df.show() // shows first n (default is 20) rows
root
|-- agency: string (nullable = true)
|-- platform: string (nullable = true)
|-- url: string (nullable = true)
|-- date: string (nullable = true)
|-- visits: integer (nullable = true)
+----------+----------+--------------------+--------------------+------+
| agency| platform| url| date|visits|
+----------+----------+--------------------+--------------------+------+
| OEM| SMS| null|02/17/2012 12:00:...| 61652|
| OEM| SMS| null|11/09/2012 12:00:...| 44547|
| EDC| Flickr|http://www.flickr...|05/09/2012 12:00:...| null|
| NYCHA|Newsletter| null|05/09/2012 12:00:...| null|
| DHS| Twitter|www.twitter.com/n...|06/13/2012 12:00:...| 389|
| DHS| Twitter|www.twitter.com/n...|08/02/2012 12:00:...| 431|
| DOH| Android| Condom Finder|08/08/2011 12:00:...| 5026|
| DOT| Android| You The Man|08/08/2011 12:00:...| null|
| MOME| Android| MiNY Venor app|08/08/2011 12:00:...| 313|
| DOT|Broadcastr| null|08/08/2011 12:00:...| null|
| DPR|Broadcastr|http://beta.broad...|08/08/2011 12:00:...| null|
| ENDHT| Facebook|http://www.facebo...|08/08/2011 12:00:...| 3|
| VAC| Facebook|https://www.faceb...|08/08/2011 12:00:...| 36|
| PlaNYC| Facebook|http://www.facebo...|08/08/2011 12:00:...| 47|
| DFTA| Facebook|http://www.facebo...|08/08/2011 12:00:...| 90|
| energyNYC| Facebook|http://www.facebo...|08/08/2011 12:00:...| 105|
| MOIA| Facebook|http://www.facebo...|08/08/2011 12:00:...| 123|
|City Store| Facebook|http://www.facebo...|08/08/2011 12:00:...| 119|
| OCDV| Facebook|http://www.facebo...|08/08/2011 12:00:...| 148|
| HIA| Facebook|http://www.facebo...|08/08/2011 12:00:...| 197|
+----------+----------+--------------------+--------------------+------+
only showing top 20 rows
Note that
(nullable = true)simply means if the value is allowed to benull.
Let us count the number of rows in df.
df.count() // Ctrl+Enter
res7: Long = 5899
So there are 5899 records or rows in the DataFrame df. Pretty good! You can also select individual columns using so-called DataFrame API, as follows:
val platforms = df.select("platform") // Shift+Enter
platforms: org.apache.spark.sql.DataFrame = [platform: string]
platforms.count() // Shift+Enter to count the number of rows
res8: Long = 5899
platforms.show(5) // Ctrl+Enter to show top 5 rows
+----------+
| platform|
+----------+
| SMS|
| SMS|
| Flickr|
|Newsletter|
| Twitter|
+----------+
only showing top 5 rows
You can also apply .distinct() to extract only unique entries as follows:
val uniquePlatforms = df.select("platform").distinct() // Shift+Enter
uniquePlatforms: org.apache.spark.sql.Dataset[org.apache.spark.sql.Row] = [platform: string]
uniquePlatforms.count() // Ctrl+Enter to count the number of distinct platforms
res10: Long = 23
Let's see all the rows of the DataFrame uniquePlatforms.
Note that
display(uniquePlatforms)unlikeuniquePlatforms.show()displays all rows of the DataFrame + gives you ability to select different view, e.g. charts.
display(uniquePlatforms) // Ctrl+Enter to show all rows; use the scroll-bar on the right of the display to see all platforms
| platform |
|---|
| nyc.gov |
| Flickr |
| Vimeo |
| iPhone |
| YouTube |
| WordPress |
| SMS |
| iPhone App |
| Youtube |
| iPhone app |
| Linked-In |
| TOTAL |
| Tumblr |
| Newsletter |
| Broadcastr |
| Android |
| Foursquare |
| Google+ |
| Foursquare (Badge Unlock) |
Spark SQL and DataFrame API
Spark SQL provides DataFrame API that can perform relational operations on both external data sources and internal collections, which is similar to widely used data frame concept in R, but evaluates operations support lazily (remember RDDs?), so that it can perform relational optimizations. This API is also available in Java, Python and R, but some functionality may not be available, although with every release of Spark people minimize this gap.
So we give some examples how to query data in Python and R, but continue with Scala. You can do all DataFrame operations in this notebook using Python or R.
# Ctrl+Enter to evaluate this python cell, recall '#' is the pre-comment character in python
# Using Python to query our "social_media_usage" table
pythonDF = spark.table("social_media_usage").select("platform").distinct()
pythonDF.show(3)
+--------+
|platform|
+--------+
| nyc.gov|
| Flickr|
| Vimeo|
+--------+
only showing top 3 rows
-- Ctrl+Enter to achieve the same result using standard SQL syntax!
select distinct platform from social_media_usage
| platform |
|---|
| nyc.gov |
| Flickr |
| Vimeo |
| iPhone |
| YouTube |
| WordPress |
| SMS |
| iPhone App |
| Youtube |
| iPhone app |
| Linked-In |
| TOTAL |
| Tumblr |
| Newsletter |
| Broadcastr |
| Android |
| Foursquare |
| Google+ |
| Foursquare (Badge Unlock) |
Now it is time for some tips around how you use select and what the difference is between $"a", col("a"), df("a").
As you probably have noticed by now, you can specify individual columns to select by providing String values in select statement. But sometimes you need to: - distinguish between columns with the same name - use it to filter (actually you can still filter using full String expression) - do some "magic" with joins and user-defined functions (this will be shown later)
So Spark gives you ability to actually specify columns when you select. Now the difference between all those three notations is ... none, those things are just aliases for a Column in Spark SQL, which means following expressions yield the same result:
// Using string expressions
df.select("agency", "visits")
// Using "$" alias for column
df.select($"agency", $"visits")
// Using "col" alias for column
df.select(col("agency"), col("visits"))
// Using DataFrame name for column
df.select(df("agency"), df("visits"))
This "same-difference" applies to filtering, i.e. you can either use full expression to filter, or column as shown in the following example:
// Using column to filter
df.select("visits").filter($"visits" > 100)
// Or you can use full expression as string
df.select("visits").filter("visits > 100")
Note that
$"visits" > 100expression looks amazing, but under the hood it is just another column, and it equals todf("visits").>(100), where, thanks to Scala paradigm>is just another function that you can define.
val sms = df.select($"agency", $"platform", $"visits").filter($"platform" === "SMS")
sms.show() // Ctrl+Enter
+------+--------+------+
|agency|platform|visits|
+------+--------+------+
| OEM| SMS| 61652|
| OEM| SMS| 44547|
| DOE| SMS| 382|
| NYCHA| SMS| null|
| OEM| SMS| 61652|
| DOE| SMS| 382|
| NYCHA| SMS| null|
| OEM| SMS| 61652|
| OEM| SMS| null|
| DOE| SMS| null|
| NYCHA| SMS| null|
| OEM| SMS| null|
| DOE| SMS| null|
| NYCHA| SMS| null|
| DOE| SMS| 382|
| NYCHA| SMS| null|
| OEM| SMS| 61652|
| DOE| SMS| 382|
| NYCHA| SMS| null|
| OEM| SMS| 61652|
+------+--------+------+
only showing top 20 rows
sms: org.apache.spark.sql.Dataset[org.apache.spark.sql.Row] = [agency: string, platform: string ... 1 more field]
Again you could have written the query above using any column aliases or String names or even writing the query directly.
For example, we can do it using String names, as follows:
// Ctrl+Enter Note that we are using "platform = 'SMS'" since it will be evaluated as actual SQL
val sms = df.select(df("agency"), df("platform"), df("visits")).filter("platform = 'SMS'")
sms.show(5)
+------+--------+------+
|agency|platform|visits|
+------+--------+------+
| OEM| SMS| 61652|
| OEM| SMS| 44547|
| DOE| SMS| 382|
| NYCHA| SMS| null|
| OEM| SMS| 61652|
+------+--------+------+
only showing top 5 rows
sms: org.apache.spark.sql.Dataset[org.apache.spark.sql.Row] = [agency: string, platform: string ... 1 more field]
Refer to the DataFrame API for more detailed API. In addition to simple column references and expressions, DataFrames also have a rich library of functions including string manipulation, date arithmetic, common math operations and more. The complete list is available in the DataFrame Function Reference.
Let's next explore some of the functionality that is available by transforming this DataFrame df into a new DataFrame called fixedDF.
- First, note that some columns are not exactly what we want them to be.
- For example date column should be standard Date/Timestamp SQL column, and
- visits should not contain null values, but
0s instead.
- Let us fix it using some code that is briefly explained here (don't worry if you don't get it completely now, you will get the hang of it by playing more)
- The
coalescefunction is similar toif-elsestatement, i.e., if first column in expression isnull, then the value of the second column is used and so on. litjust means column of constant value (literally speaking!).- the "funky" time conversion is essentially conversion from current format -> unix timestamp as a number -> Spark SQL Date format
- we also remove
TOTALvalue fromplatformcolumn.
- The
// Ctrl+Enter to make fixedDF
// import the needed sql functions
import org.apache.spark.sql.functions.{coalesce, from_unixtime, lit, to_date, unix_timestamp}
// make the fixedDF DataFrame
val fixedDF = df.
select(
$"agency",
$"platform",
$"url",
to_date(from_unixtime(unix_timestamp($"date", "MM/dd/yyyy hh:mm:ss aaa"))).as("date"),
coalesce($"visits", lit(0)).as("visits")).
filter($"platform" !== "TOTAL")
fixedDF.printSchema() // print its schema
// and show the top 20 records fully
fixedDF.show(false) // the false argument does not truncate the rows, so you will not see something like this "anot..."
root
|-- agency: string (nullable = true)
|-- platform: string (nullable = true)
|-- url: string (nullable = true)
|-- date: date (nullable = true)
|-- visits: integer (nullable = false)
+----------+----------+---------------------------------------------------------------------------------------+----------+------+
|agency |platform |url |date |visits|
+----------+----------+---------------------------------------------------------------------------------------+----------+------+
|OEM |SMS |null |2012-02-17|61652 |
|OEM |SMS |null |2012-11-09|44547 |
|EDC |Flickr |http://www.flickr.com/nycedc |2012-05-09|0 |
|NYCHA |Newsletter|null |2012-05-09|0 |
|DHS |Twitter |www.twitter.com/nycdhs |2012-06-13|389 |
|DHS |Twitter |www.twitter.com/nycdhs |2012-08-02|431 |
|DOH |Android |Condom Finder |2011-08-08|5026 |
|DOT |Android |You The Man |2011-08-08|0 |
|MOME |Android |MiNY Venor app |2011-08-08|313 |
|DOT |Broadcastr|null |2011-08-08|0 |
|DPR |Broadcastr|http://beta.broadcastr.com/Echo.html?audioId=670026-4001 |2011-08-08|0 |
|ENDHT |Facebook |http://www.facebook.com/pages/NYC-Lets-End-Human-Trafficking/125730490795659?sk=wall |2011-08-08|3 |
|VAC |Facebook |https://www.facebook.com/pages/NYC-Voter-Assistance-Commission/110226709012110 |2011-08-08|36 |
|PlaNYC |Facebook |http://www.facebook.com/pages/New-York-NY/PlaNYC/160454173971169?ref=ts |2011-08-08|47 |
|DFTA |Facebook |http://www.facebook.com/pages/NYC-Department-for-the-Aging/109028655823590 |2011-08-08|90 |
|energyNYC |Facebook |http://www.facebook.com/EnergyNYC?sk=wall |2011-08-08|105 |
|MOIA |Facebook |http://www.facebook.com/ihwnyc |2011-08-08|123 |
|City Store|Facebook |http://www.facebook.com/citystorenyc |2011-08-08|119 |
|OCDV |Facebook |http://www.facebook.com/pages/NYC-Healthy-Relationship-Training-Academy/134637829901065|2011-08-08|148 |
|HIA |Facebook |http://www.facebook.com/pages/New-York-City-Health-Insurance-Link/145920551598 |2011-08-08|197 |
+----------+----------+---------------------------------------------------------------------------------------+----------+------+
only showing top 20 rows
<console>:47: warning: method !== in class Column is deprecated: !== does not have the same precedence as ===, use =!= instead
filter($"platform" !== "TOTAL")
^
import org.apache.spark.sql.functions.{coalesce, from_unixtime, lit, to_date, unix_timestamp}
fixedDF: org.apache.spark.sql.Dataset[org.apache.spark.sql.Row] = [agency: string, platform: string ... 3 more fields]
Okay, this is better, but urls are still inconsistent.
Let's fix this by writing our own UDF (user-defined function) to deal with special cases.
Note that if you CAN USE Spark functions library, do it. But for the sake of the example, custom UDF is shown below.
We take value of a column as String type and return the same String type, but ignore values that do not start with http.
// Ctrl+Enter to evaluate this UDF which takes a input String called "value"
// and converts it into lower-case if it begins with http and otherwise leaves it as null, so we sort of remove non valid web-urls
val cleanUrl = udf((value: String) => if (value != null && value.startsWith("http")) value.toLowerCase() else null)
cleanUrl: org.apache.spark.sql.expressions.UserDefinedFunction = UserDefinedFunction(<function1>,StringType,Some(List(StringType)))
Let us apply our UDF on fixedDF to create a new DataFrame called cleanedDF as follows:
// Ctrl+Enter
val cleanedDF = fixedDF.select($"agency", $"platform", cleanUrl($"url").as("url"), $"date", $"visits")
cleanedDF: org.apache.spark.sql.DataFrame = [agency: string, platform: string ... 3 more fields]
Now, let's check that it actually worked by seeing the first 5 rows of the cleanedDF whose url isNull and isNotNull, as follows:
// Shift+Enter
cleanedDF.filter($"url".isNull).show(5)
+------+----------+----+----------+------+
|agency| platform| url| date|visits|
+------+----------+----+----------+------+
| OEM| SMS|null|2012-02-17| 61652|
| OEM| SMS|null|2012-11-09| 44547|
| NYCHA|Newsletter|null|2012-05-09| 0|
| DHS| Twitter|null|2012-06-13| 389|
| DHS| Twitter|null|2012-08-02| 431|
+------+----------+----+----------+------+
only showing top 5 rows
// Ctrl+Enter
cleanedDF.filter($"url".isNotNull).show(5, false) // false in .show(5, false) shows rows untruncated
+------+----------+------------------------------------------------------------------------------------+----------+------+
|agency|platform |url |date |visits|
+------+----------+------------------------------------------------------------------------------------+----------+------+
|EDC |Flickr |http://www.flickr.com/nycedc |2012-05-09|0 |
|DPR |Broadcastr|http://beta.broadcastr.com/echo.html?audioid=670026-4001 |2011-08-08|0 |
|ENDHT |Facebook |http://www.facebook.com/pages/nyc-lets-end-human-trafficking/125730490795659?sk=wall|2011-08-08|3 |
|VAC |Facebook |https://www.facebook.com/pages/nyc-voter-assistance-commission/110226709012110 |2011-08-08|36 |
|PlaNYC|Facebook |http://www.facebook.com/pages/new-york-ny/planyc/160454173971169?ref=ts |2011-08-08|47 |
+------+----------+------------------------------------------------------------------------------------+----------+------+
only showing top 5 rows
Now there is a suggestion from you manager's manager's manager that due to some perceived privacy concerns we want to replace agency with some unique identifier.
So we need to do the following: * create unique list of agencies with ids and * join them with main DataFrame.
Sounds easy, right? Let's do it.
// Crtl+Enter
// Import Spark SQL function that will give us unique id across all the records in this DataFrame
import org.apache.spark.sql.functions.monotonically_increasing_id
// We append column as SQL function that creates unique ids across all records in DataFrames
val agencies = cleanedDF.select(cleanedDF("agency"))
.distinct()
.withColumn("id", monotonically_increasing_id())
agencies.show(5)
+--------------------+-----------+
| agency| id|
+--------------------+-----------+
| PlaNYC|34359738368|
| HIA|34359738369|
|NYC Digital: exte...|34359738370|
| NYCGLOBAL|42949672960|
| nycgov|68719476736|
+--------------------+-----------+
only showing top 5 rows
import org.apache.spark.sql.functions.monotonically_increasing_id
agencies: org.apache.spark.sql.DataFrame = [agency: string, id: bigint]
Those who want to understand left/right inner/outer joins can see the video lectures in Module 3 of Anthony Joseph's Introduction to Big data edX course from the Community Edition of databricks. The course has been added to this databricks shard at /#workspace/scalable-data-science/xtraResources/edXBigDataSeries2015/CS100-1x as extra resources for the project-focussed course Scalable Data Science.
// Ctrl+Enter
// And join with the rest of the data, note how join condition is specified
val anonym = cleanedDF.join(agencies, cleanedDF("agency") === agencies("agency"), "inner").select("id", "platform", "url", "date", "visits")
// We also cache DataFrame since it can be quite expensive to recompute join
anonym.cache()
// Display result
anonym.show(5)
+-------------+----------+--------------------+----------+------+
| id| platform| url| date|visits|
+-------------+----------+--------------------+----------+------+
|1580547964928| SMS| null|2012-02-17| 61652|
|1580547964928| SMS| null|2012-11-09| 44547|
| 412316860416| Flickr|http://www.flickr...|2012-05-09| 0|
|1649267441664|Newsletter| null|2012-05-09| 0|
|1529008357376| Twitter| null|2012-06-13| 389|
+-------------+----------+--------------------+----------+------+
only showing top 5 rows
anonym: org.apache.spark.sql.DataFrame = [id: bigint, platform: string ... 3 more fields]
spark.catalog.listTables().show() // look at the available tables
+--------------------+--------+-----------+---------+-----------+
| name|database|description|tableType|isTemporary|
+--------------------+--------+-----------+---------+-----------+
| cities_csv| default| null| EXTERNAL| false|
| cleaned_taxes| default| null| MANAGED| false|
|commdettrumpclint...| default| null| MANAGED| false|
| donaldtrumptweets| default| null| EXTERNAL| false|
| linkage| default| null| EXTERNAL| false|
| nations| default| null| EXTERNAL| false|
| newmplist| default| null| EXTERNAL| false|
| ny_baby_names| default| null| MANAGED| false|
| nzmpsandparty| default| null| EXTERNAL| false|
| pos_neg_category| default| null| EXTERNAL| false|
| rna| default| null| MANAGED| false|
| samh| default| null| EXTERNAL| false|
| social_media_usage| default| null| EXTERNAL| false|
| table1| default| null| EXTERNAL| false|
| test_table| default| null| EXTERNAL| false|
| uscites| default| null| EXTERNAL| false|
+--------------------+--------+-----------+---------+-----------+
-- to remove a TempTable if it exists already
drop table if exists anonym
// Register table for Spark SQL, we also import "month" function
import org.apache.spark.sql.functions.month
anonym.createOrReplaceTempView("anonym")
import org.apache.spark.sql.functions.month
-- Interesting. Now let's do some aggregation. Display platform, month, visits
-- Date column allows us to extract month directly
select platform, month(date) as month, sum(visits) as visits from anonym group by platform, month(date)
| platform | month | visits |
|---|---|---|
| 9.0 | 27891.0 | |
| Linked-In | 10.0 | 60156.0 |
| Foursquare (Badge Unlock) | 6.0 | 0.0 |
| iPhone | 8.0 | 10336.0 |
| 1.0 | 0.0 | |
| 9.0 | 819290.0 | |
| Vimeo | 11.0 | 0.0 |
| Linked-In | 1.0 | 19007.0 |
| iPhone app | 9.0 | 33348.0 |
| SMS | 10.0 | 54100.0 |
| YouTube | 2.0 | 4937.0 |
| 11.0 | 58968.0 | |
| YouTube | 3.0 | 6066.0 |
| iPhone | 2.0 | 0.0 |
| Newsletter | 11.0 | 3079091.0 |
| Google+ | 2.0 | 0.0 |
| Android | 4.0 | 724.0 |
| 2.0 | 0.0 | |
| Android | 3.0 | 343.0 |
| Youtube | 10.0 | 429.0 |
| Android | 11.0 | 11259.0 |
| Newsletter | 12.0 | 1606654.0 |
| iPhone App | 4.0 | 55960.0 |
| iPhone app | 12.0 | 21352.0 |
| 3.0 | 291971.0 | |
| Google+ | 5.0 | 0.0 |
| Newsletter | 5.0 | 847813.0 |
| 10.0 | 28145.0 | |
| Linked-In | 7.0 | 31758.0 |
| Tumblr | 5.0 | 26932.0 |
| SMS | 2.0 | 62034.0 |
| Linked-In | 6.0 | 30867.0 |
| YouTube | 11.0 | 22820.0 |
| Foursquare | 3.0 | 25786.0 |
| SMS | 11.0 | 170234.0 |
| Foursquare (Badge Unlock) | 11.0 | 22512.0 |
| Youtube | 11.0 | 770.0 |
| iPhone App | 2.0 | 21672.0 |
| YouTube | 12.0 | 9505.0 |
| Foursquare | 8.0 | 42230.0 |
| Foursquare (Badge Unlock) | 4.0 | 20152.0 |
| 5.0 | 351601.0 | |
| Tumblr | 3.0 | 5098.0 |
| Linked-In | 8.0 | 45346.0 |
| Google+ | 10.0 | 8.0 |
| 6.0 | 399330.0 | |
| Foursquare | 1.0 | 10126.0 |
| YouTube | 4.0 | 12542.0 |
| Newsletter | 6.0 | 1137677.0 |
| YouTube | 7.0 | 6748.0 |
| Google+ | 4.0 | 0.0 |
| Youtube | 6.0 | 229.0 |
| Linked-In | 11.0 | 75747.0 |
| 1.0 | 259797.0 | |
| Youtube | 5.0 | 0.0 |
| Vimeo | 10.0 | 0.0 |
| iPhone App | 1.0 | 21672.0 |
| Android | 5.0 | 381.0 |
| Flickr | 8.0 | 493.0 |
| iPhone app | 8.0 | 41389.0 |
| 10.0 | 1010914.0 | |
| Foursquare (Badge Unlock) | 10.0 | 11256.0 |
| 8.0 | 5995.0 | |
| Vimeo | 12.0 | 0.0 |
| Flickr | 1.0 | 217.0 |
| WordPress | 2.0 | 0.0 |
| Android | 10.0 | 5473.0 |
| Youtube | 2.0 | 155.0 |
| Android | 6.0 | 102.0 |
| Tumblr | 7.0 | 47121.0 |
| 1.0 | 0.0 | |
| Linked-In | 3.0 | 20761.0 |
| iPhone app | 10.0 | 30713.0 |
| Youtube | 3.0 | 163.0 |
| WordPress | 6.0 | 4641.0 |
| iPhone | 11.0 | 25848.0 |
| Broadcastr | 3.0 | 0.0 |
| Youtube | 7.0 | 240.0 |
| iPhone App | 11.0 | 96342.0 |
| SMS | 12.0 | 124068.0 |
| 4.0 | 674303.0 | |
| Youtube | 12.0 | 291.0 |
| 1.0 | 364376.0 | |
| Newsletter | 3.0 | 803327.0 |
| iPhone | 5.0 | 8203.0 |
| iPhone app | 5.0 | 30598.0 |
| Tumblr | 8.0 | 54224.0 |
| iPhone App | 12.0 | 43344.0 |
| WordPress | 1.0 | 0.0 |
| iPhone app | 3.0 | 10676.0 |
| Flickr | 12.0 | 432.0 |
| 10.0 | 859354.0 | |
| Android | 12.0 | 686.0 |
| Vimeo | 6.0 | 0.0 |
| Vimeo | 8.0 | 0.0 |
| iPhone App | 6.0 | 34966.0 |
| iPhone | 6.0 | 9643.0 |
| SMS | 5.0 | 0.0 |
| Vimeo | 7.0 | 0.0 |
| Android | 1.0 | 343.0 |
| WordPress | 12.0 | 0.0 |
| iPhone app | 6.0 | 2713.0 |
| Newsletter | 10.0 | 2359712.0 |
| 11.0 | 312.0 | |
| Broadcastr | 6.0 | 0.0 |
| Google+ | 1.0 | 0.0 |
| Android | 8.0 | 5784.0 |
| 6.0 | 0.0 | |
| Newsletter | 4.0 | 1606654.0 |
| Newsletter | 9.0 | 1941202.0 |
| WordPress | 10.0 | 66299.0 |
| iPhone | 1.0 | 0.0 |
| SMS | 8.0 | 116134.0 |
| Foursquare (Badge Unlock) | 12.0 | 0.0 |
| 7.0 | 470477.0 | |
| Linked-In | 12.0 | 35231.0 |
| Foursquare (Badge Unlock) | 1.0 | 0.0 |
| iPhone | 4.0 | 8203.0 |
| Flickr | 3.0 | 227.0 |
| iPhone app | 11.0 | 72102.0 |
| YouTube | 1.0 | 4904.0 |
| Flickr | 10.0 | 153231.0 |
| 4.0 | 3404.0 | |
| WordPress | 8.0 | 5017.0 |
| YouTube | 9.0 | 12107.0 |
| iPhone app | 4.0 | 41274.0 |
| Broadcastr | 12.0 | 0.0 |
| Linked-In | 4.0 | 43582.0 |
| YouTube | 8.0 | 10974.0 |
| iPhone | 12.0 | 0.0 |
| Newsletter | 1.0 | 803327.0 |
| Broadcastr | 9.0 | 0.0 |
| Broadcastr | 2.0 | 0.0 |
| Flickr | 5.0 | 287.0 |
| Tumblr | 10.0 | 97401.0 |
| Linked-In | 2.0 | 19920.0 |
| Broadcastr | 8.0 | 0.0 |
| iPhone App | 9.0 | 42128.0 |
| 2.0 | 0.0 | |
| 4.0 | 844718.0 | |
| Broadcastr | 10.0 | 0.0 |
| Foursquare | 12.0 | 19110.0 |
| Tumblr | 6.0 | 41248.0 |
| Vimeo | 2.0 | 0.0 |
| WordPress | 4.0 | 0.0 |
| 11.0 | 1660542.0 | |
| Linked-In | 9.0 | 49299.0 |
| 12.0 | 690189.0 | |
| Android | 9.0 | 445.0 |
| iPhone | 9.0 | 12924.0 |
| WordPress | 9.0 | 4897.0 |
| Tumblr | 1.0 | 2645.0 |
| Foursquare | 2.0 | 21181.0 |
| Foursquare | 10.0 | 69598.0 |
| Broadcastr | 1.0 | 0.0 |
| 10.0 | 141.0 | |
| 9.0 | 754875.0 | |
| YouTube | 10.0 | 9100.0 |
| 3.0 | 400250.0 | |
| Broadcastr | 11.0 | 0.0 |
| 8.0 | 38.0 | |
| 6.0 | 461261.0 | |
| Foursquare (Badge Unlock) | 2.0 | 8878.0 |
| Google+ | 12.0 | 0.0 |
| Google+ | 3.0 | 0.0 |
| Linked-In | 5.0 | 29808.0 |
| Vimeo | 1.0 | 0.0 |
| Foursquare | 5.0 | 29991.0 |
| Foursquare | 7.0 | 38590.0 |
| iPhone app | 7.0 | 30713.0 |
| Newsletter | 2.0 | 803327.0 |
| 8.0 | 704438.0 | |
| 2.0 | 107993.0 | |
| Broadcastr | 4.0 | 0.0 |
| 5.0 | 0.0 | |
| Android | 7.0 | 445.0 |
| Flickr | 9.0 | 549.0 |
| WordPress | 11.0 | 9294.0 |
| Flickr | 11.0 | 1007.0 |
| 4.0 | 0.0 | |
| iPhone | 10.0 | 12924.0 |
| 12.0 | 0.0 | |
| Flickr | 4.0 | 545.0 |
| 7.0 | 451076.0 | |
| Google+ | 8.0 | 0.0 |
| Tumblr | 9.0 | 62742.0 |
| Android | 2.0 | 343.0 |
| Foursquare | 4.0 | 57337.0 |
| SMS | 1.0 | 62034.0 |
| WordPress | 3.0 | 0.0 |
| 3.0 | 0.0 | |
| iPhone App | 3.0 | 21672.0 |
| Tumblr | 4.0 | 31247.0 |
| 9.0 | 74.0 | |
| Broadcastr | 5.0 | 0.0 |
| YouTube | 5.0 | 0.0 |
| 8.0 | 657312.0 | |
| iPhone App | 5.0 | 34288.0 |
| Youtube | 1.0 | 150.0 |
| SMS | 9.0 | 116134.0 |
| iPhone | 3.0 | 0.0 |
| Vimeo | 5.0 | 0.0 |
| Google+ | 7.0 | 0.0 |
| SMS | 6.0 | 54100.0 |
| Vimeo | 9.0 | 0.0 |
| Google+ | 11.0 | 26.0 |
| 7.0 | 0.0 | |
| Foursquare (Badge Unlock) | 9.0 | 11256.0 |
| SMS | 3.0 | 62034.0 |
| 7.0 | 5450.0 | |
| iPhone App | 10.0 | 64589.0 |
| 12.0 | 0.0 | |
| Flickr | 2.0 | 219.0 |
| 6.0 | 4764.0 | |
| Foursquare (Badge Unlock) | 7.0 | 0.0 |
| Flickr | 6.0 | 332.0 |
| SMS | 7.0 | 54100.0 |
| Tumblr | 12.0 | 5005.0 |
| 5.0 | 435148.0 | |
| Youtube | 9.0 | 281.0 |
| iPhone App | 8.0 | 57513.0 |
| Google+ | 9.0 | 0.0 |
| Foursquare (Badge Unlock) | 8.0 | 11256.0 |
| 5.0 | 0.0 | |
| 11.0 | 1408965.0 | |
| 2.0 | 385091.0 | |
| iPhone App | 7.0 | 35841.0 |
| iPhone app | 1.0 | 10676.0 |
| Vimeo | 3.0 | 0.0 |
| SMS | 4.0 | 124068.0 |
| WordPress | 7.0 | 4647.0 |
| Youtube | 4.0 | 367.0 |
| Foursquare (Badge Unlock) | 5.0 | 0.0 |
| Foursquare | 6.0 | 34193.0 |
| Google+ | 6.0 | 0.0 |
| Youtube | 8.0 | 258.0 |
| 3.0 | 0.0 | |
| Newsletter | 7.0 | 1137868.0 |
| Foursquare | 9.0 | 50489.0 |
| Foursquare | 11.0 | 118323.0 |
| iPhone app | 2.0 | 10676.0 |
| Flickr | 7.0 | 342.0 |
| Newsletter | 8.0 | 1941197.0 |
| Tumblr | 11.0 | 195881.0 |
| Foursquare (Badge Unlock) | 3.0 | 0.0 |
| 12.0 | 502687.0 | |
| Broadcastr | 7.0 | 0.0 |
| WordPress | 5.0 | 0.0 |
| YouTube | 6.0 | 6509.0 |
| Tumblr | 2.0 | 4406.0 |
| Vimeo | 4.0 | 0.0 |
| iPhone | 7.0 | 10336.0 |
Note, that we could have done aggregation using DataFrame API instead of Spark SQL.
Alright, now let's see some cool operations with window functions.
Our next task is to compute (daily visits / monthly average) for all platforms.
import org.apache.spark.sql.functions.{dayofmonth, month, row_number, sum}
import org.apache.spark.sql.expressions.Window
val coolDF = anonym.select($"id", $"platform", dayofmonth($"date").as("day"), month($"date").as("month"), $"visits").
groupBy($"id", $"platform", $"day", $"month").agg(sum("visits").as("visits"))
// Run window aggregation on visits per month and platform
val window = coolDF.select($"id", $"day", $"visits", sum($"visits").over(Window.partitionBy("platform", "month")).as("monthly_visits"))
// Create and register percent table
val percent = window.select($"id", $"day", ($"visits" / $"monthly_visits").as("percent"))
percent.createOrReplaceTempView("percent")
import org.apache.spark.sql.functions.{dayofmonth, month, row_number, sum}
import org.apache.spark.sql.expressions.Window
coolDF: org.apache.spark.sql.DataFrame = [id: bigint, platform: string ... 3 more fields]
window: org.apache.spark.sql.DataFrame = [id: bigint, day: int ... 2 more fields]
percent: org.apache.spark.sql.DataFrame = [id: bigint, day: int ... 1 more field]
-- A little bit of visualization as result of our efforts
select id, day, `percent` from percent where `percent` > 0.3 and day = 2
| id | day | percent |
|---|---|---|
| 6.52835028992e11 | 2.0 | 1.0 |
| 5.06806140929e11 | 2.0 | 0.4993894993894994 |
| 6.52835028992e11 | 2.0 | 0.446576072475353 |
| 9.01943132161e11 | 2.0 | 0.5 |
| 6.52835028992e11 | 2.0 | 0.3181818181818182 |
| 9.01943132161e11 | 2.0 | 0.6180914042150131 |
| 2.147483648e11 | 2.0 | 0.3663035756571158 |
| 1.322849927168e12 | 2.0 | 0.5265514047545539 |
| 1.322849927168e12 | 2.0 | 0.3109034021149352 |
| 1.408749273089e12 | 2.0 | 0.6937119675456389 |
| 6.52835028992e11 | 2.0 | 0.6765082509845611 |
| 5.06806140929e11 | 2.0 | 1.0 |
| 6.52835028992e11 | 2.0 | 0.5 |
| 4.12316860416e11 | 2.0 | 0.3408084980820301 |
| 1.580547964928e12 | 2.0 | 0.383582757848692 |
| 6.52835028992e11 | 2.0 | 0.38833874233724447 |
| 2.06158430208e11 | 2.0 | 0.9262507474586407 |
| 1.666447310848e12 | 2.0 | 0.9473684210526315 |
| 2.06158430208e11 | 2.0 | 0.5 |
| 1.408749273089e12 | 2.0 | 0.394240317775571 |
| 6.8719476736e10 | 2.0 | 0.38461538461538464 |
| 1.640677507072e12 | 2.0 | 0.44748143897901344 |
| 9.01943132161e11 | 2.0 | 1.0 |
| 6.52835028992e11 | 2.0 | 0.8449612403100775 |
| 9.01943132161e11 | 2.0 | 0.3060168545490231 |
-- You also could just use plain SQL to write query above, note that you might need to group by id and day as well.
with aggr as (
select id, dayofmonth(date) as day, visits / sum(visits) over (partition by (platform, month(date))) as percent
from anonym
)
select * from aggr where day = 2 and percent > 0.3
| id | day | percent |
|---|---|---|
| 6.52835028992e11 | 2.0 | 1.0 |
| 5.06806140929e11 | 2.0 | 0.4993894993894994 |
| 6.52835028992e11 | 2.0 | 0.446576072475353 |
| 9.01943132161e11 | 2.0 | 0.5 |
| 6.52835028992e11 | 2.0 | 0.3181818181818182 |
| 2.147483648e11 | 2.0 | 0.3663035756571158 |
| 9.01943132161e11 | 2.0 | 0.6180914042150131 |
| 1.322849927168e12 | 2.0 | 0.4718608035989944 |
| 1.408749273089e12 | 2.0 | 0.6937119675456389 |
| 6.52835028992e11 | 2.0 | 0.6765082509845611 |
| 5.06806140929e11 | 2.0 | 1.0 |
| 6.52835028992e11 | 2.0 | 0.5 |
| 4.12316860416e11 | 2.0 | 0.3408084980820301 |
| 1.580547964928e12 | 2.0 | 0.383582757848692 |
| 6.52835028992e11 | 2.0 | 0.38833874233724447 |
| 2.06158430208e11 | 2.0 | 0.9262507474586407 |
| 1.666447310848e12 | 2.0 | 0.9473684210526315 |
| 2.06158430208e11 | 2.0 | 0.5 |
| 1.408749273089e12 | 2.0 | 0.394240317775571 |
| 6.8719476736e10 | 2.0 | 0.38461538461538464 |
| 1.640677507072e12 | 2.0 | 0.44748143897901344 |
| 9.01943132161e11 | 2.0 | 1.0 |
| 6.52835028992e11 | 2.0 | 0.8449612403100775 |
| 9.01943132161e11 | 2.0 | 0.3060168545490231 |
Interoperating with RDDs
Spark SQL supports two different methods for converting existing RDDs into DataFrames. The first method uses reflection to infer the schema of an RDD that contains specific types of objects. This reflection based approach leads to more concise code and works well when you already know the schema.
The second method for creating DataFrames is through a programmatic interface that allows you to construct a schema and then apply it to an existing RDD. While this method is more verbose, it allows you to construct DataFrames when the columns and their types are not known until runtime.
Inferring the Schema Using Reflection
The Scala interface for Spark SQL supports automatically converting an RDD containing case classes to a DataFrame. The case class defines the schema of the table. The names of the arguments to the case class are read using reflection and become the names of the columns. Case classes can also be nested or contain complex types such as Sequences or Arrays. This RDD can be implicitly converted to a DataFrame and then be registered as a table.
// Define case class that will be our schema for DataFrame
case class Hubot(name: String, year: Int, manufacturer: String, version: Array[Int], details: Map[String, String])
// You can process a text file, for example, to convert every row to our Hubot, but we will create RDD manually
val rdd = sc.parallelize(
Array(
Hubot("Jerry", 2015, "LCorp", Array(1, 2, 3), Map("eat" -> "yes", "sleep" -> "yes", "drink" -> "yes")),
Hubot("Mozart", 2010, "LCorp", Array(1, 2), Map("eat" -> "no", "sleep" -> "no", "drink" -> "no")),
Hubot("Einstein", 2012, "LCorp", Array(1, 2, 3), Map("eat" -> "yes", "sleep" -> "yes", "drink" -> "no"))
)
)
defined class Hubot
rdd: org.apache.spark.rdd.RDD[Hubot] = ParallelCollectionRDD[23107] at parallelize at <console>:45
// In order to convert RDD into DataFrame you need to do this:
val hubots = rdd.toDF()
// Display DataFrame, note how array and map fields are displayed
hubots.printSchema()
hubots.show()
root
|-- name: string (nullable = true)
|-- year: integer (nullable = false)
|-- manufacturer: string (nullable = true)
|-- version: array (nullable = true)
| |-- element: integer (containsNull = false)
|-- details: map (nullable = true)
| |-- key: string
| |-- value: string (valueContainsNull = true)
+--------+----+------------+---------+--------------------+
| name|year|manufacturer| version| details|
+--------+----+------------+---------+--------------------+
| Jerry|2015| LCorp|[1, 2, 3]|Map(eat -> yes, s...|
| Mozart|2010| LCorp| [1, 2]|Map(eat -> no, sl...|
|Einstein|2012| LCorp|[1, 2, 3]|Map(eat -> yes, s...|
+--------+----+------------+---------+--------------------+
hubots: org.apache.spark.sql.DataFrame = [name: string, year: int ... 3 more fields]
// You can query complex type the same as you query any other column
// By the way you can use `sql` function to invoke Spark SQL to create DataFrame
hubots.createOrReplaceTempView("hubots")
val onesThatEat = sqlContext.sql("select name, details.eat from hubots where details.eat = 'yes'")
onesThatEat.show()
+--------+---+
| name|eat|
+--------+---+
| Jerry|yes|
|Einstein|yes|
+--------+---+
onesThatEat: org.apache.spark.sql.DataFrame = [name: string, eat: string]
Programmatically Specifying the Schema
When case classes cannot be defined ahead of time (for example, the structure of records is encoded in a string, or a text dataset will be parsed and fields will be projected differently for different users), a DataFrame can be created programmatically with three steps.
- Create an RDD of
Rows from the original RDD - Create the schema represented by a StructType and StructField classes matching the structure of
Rows in the RDD created in Step 1. - Apply the schema to the RDD of
Rows viacreateDataFramemethod provided bySQLContext.
import org.apache.spark.sql.types._
// Let's say we have an RDD of String and we need to convert it into a DataFrame with schema "name", "year", and "manufacturer"
// As you can see every record is space-separated.
val rdd = sc.parallelize(
Array(
"Jerry 2015 LCorp",
"Mozart 2010 LCorp",
"Einstein 2012 LCorp"
)
)
// Create schema as StructType //
val schema = StructType(
StructField("name", StringType, false) ::
StructField("year", IntegerType, false) ::
StructField("manufacturer", StringType, false) ::
Nil
)
// Prepare RDD[Row]
val rows = rdd.map { entry =>
val arr = entry.split("\\s+")
val name = arr(0)
val year = arr(1).toInt
val manufacturer = arr(2)
Row(name, year, manufacturer)
}
// Create DataFrame
val bots = sqlContext.createDataFrame(rows, schema)
bots.printSchema()
bots.show()
root
|-- name: string (nullable = false)
|-- year: integer (nullable = false)
|-- manufacturer: string (nullable = false)
+--------+----+------------+
| name|year|manufacturer|
+--------+----+------------+
| Jerry|2015| LCorp|
| Mozart|2010| LCorp|
|Einstein|2012| LCorp|
+--------+----+------------+
import org.apache.spark.sql.types._
rdd: org.apache.spark.rdd.RDD[String] = ParallelCollectionRDD[23118] at parallelize at <console>:47
schema: org.apache.spark.sql.types.StructType = StructType(StructField(name,StringType,false), StructField(year,IntegerType,false), StructField(manufacturer,StringType,false))
rows: org.apache.spark.rdd.RDD[org.apache.spark.sql.Row] = MapPartitionsRDD[23119] at map at <console>:64
bots: org.apache.spark.sql.DataFrame = [name: string, year: int ... 1 more field]
Creating Datasets
A Dataset is a strongly-typed, immutable collection of objects that are mapped to a relational schema. At the core of the Dataset API is a new concept called an encoder, which is responsible for converting between JVM objects and tabular representation. The tabular representation is stored using Spark’s internal Tungsten binary format, allowing for operations on serialized data and improved memory utilization. Spark 2.2 comes with support for automatically generating encoders for a wide variety of types, including primitive types (e.g. String, Integer, Long), and Scala case classes.
Simply put, you will get all the benefits of DataFrames with fair amount of flexibility of RDD API.
// We can start working with Datasets by using our "hubots" table
// To create Dataset from DataFrame do this (assuming that case class Hubot exists):
val ds = hubots.as[Hubot]
ds.show()
+--------+----+------------+---------+--------------------+
| name|year|manufacturer| version| details|
+--------+----+------------+---------+--------------------+
| Jerry|2015| LCorp|[1, 2, 3]|Map(eat -> yes, s...|
| Mozart|2010| LCorp| [1, 2]|Map(eat -> no, sl...|
|Einstein|2012| LCorp|[1, 2, 3]|Map(eat -> yes, s...|
+--------+----+------------+---------+--------------------+
ds: org.apache.spark.sql.Dataset[Hubot] = [name: string, year: int ... 3 more fields]
Side-note: Dataset API is first-class citizen in Spark, and DataFrame is an alias for Dataset[Row]. Note that Python and R still use DataFrames (since they are dynamically typed), but it is essentially a Dataset.
Finally
DataFrames and Datasets can simplify and improve most of the applications pipelines by bringing concise syntax and performance optimizations. We would highly recommend you to check out the official API documentation, specifically around * DataFrame API, * Spark SQL functions library, * GroupBy clause and aggregated functions.
Unfortunately, this is just a getting started quickly course, and we skip features like custom aggregations, types, pivoting, etc., but if you are keen to know then start from the links above and this notebook and others in this directory.
Appendix
How to download data and make a table
Okay, so how did we actually make table "socialmediausage"? Databricks allows us to upload/link external data and make it available as registerd SQL table. It involves several steps: 1. Find interesting set of data - Google can be your friend for most cases here, or you can have your own dataset as CSV file, for example. Good source of data can also be found here: http://www.data.gov/ 2. Download / prepare it to be either on S3, or human-readable format like CSV, or JSON 3. Go to Databricks cloud (where you log in to use Databricks notebooks) and open tab Tables 4. On the very top of the left sub-menu you will see button + Create table, click on it 5. You will see page with drop-down menu of the list of sources you can provide, File means any file (Parquet, Avro, CSV), but it works the best with CSV format 6. Once you have chosen file and loaded it, you can change column names, or tweak types (mainly for CSV format) 7. That is it. Just click on final button to create table. After that you can refer to the table using sqlContext.table("YOUR_TABLE_NAME")
This is an elaboration of the Apache Spark 2.2 sql-progamming-guide.
Data Sources
Spark Sql Programming Guide
- Overview
- SQL
- DataFrames
- Datasets
- Getting Started
- Starting Point: SQLContext
- Creating DataFrames
- DataFrame Operations
- Running SQL Queries Programmatically
- Creating Datasets
- Interoperating with RDDs
- Inferring the Schema Using Reflection
- Programmatically Specifying the Schema
- Data Sources
- Generic Load/Save Functions
- Manually Specifying Options
- Run SQL on files directly
- Save Modes
- Saving to Persistent Tables
- Parquet Files
- Loading Data Programmatically
- Partition Discovery
- Schema Merging
- Hive metastore Parquet table conversion
- Hive/Parquet Schema Reconciliation
- Metadata Refreshing
- Configuration
- JSON Datasets
- Hive Tables
- Interacting with Different Versions of Hive Metastore
- JDBC To Other Databases
- Troubleshooting
- Generic Load/Save Functions
- Performance Tuning
- Caching Data In Memory
- Other Configuration Options
- Distributed SQL Engine
- Running the Thrift JDBC/ODBC server
- Running the Spark SQL CLI
Data Sources
Spark SQL supports operating on a variety of data sources through the DataFrame or DataFrame interfaces. A Dataset can be operated on as normal RDDs and can also be registered as a temporary table. Registering a Dataset as a table allows you to run SQL queries over its data. But from time to time you would need to either load or save Dataset. Spark SQL provides built-in data sources as well as Data Source API to define your own data source and use it read / write data into Spark.
Overview
Spark provides some built-in datasources that you can use straight out of the box, such as Parquet, JSON, JDBC, ORC (available with enabled Hive Support, but this is changing, and ORC will not require Hive support and will work with default Spark session starting from next release), and Text (since Spark 1.6) and CSV (since Spark 2.0, before that it is accessible as a package).
Third-party datasource packages
Community also have built quite a few datasource packages to provide easy access to the data from other formats. You can find list of those packages on http://spark-packages.org/, e.g. Avro, CSV, Amazon Redshit (for Spark < 2.0), XML, NetFlow and many others.
Generic Load/Save functions
In order to load or save DataFrame you have to call either read or write. This will return DataFrameReader or DataFrameWriter depending on what you are trying to achieve. Essentially these classes are entry points to the reading / writing actions. They allow you to specify writing mode or provide additional options to read data source.
// This will return DataFrameReader to read data source
println(spark.read)
val df = spark.range(0, 10)
// This will return DataFrameWriter to save DataFrame
println(df.write)
org.apache.spark.sql.DataFrameReader@7e7dee07
org.apache.spark.sql.DataFrameWriter@1f45f4b
df: org.apache.spark.sql.Dataset[Long] = [id: bigint]
// Saving Parquet table in Scala
val df_save = spark.table("social_media_usage").select("platform", "visits")
df_save.write.mode("overwrite").parquet("/tmp/platforms.parquet")
// Loading Parquet table in Scala
val df = spark.read.parquet("/tmp/platforms.parquet")
df.show(5)
+----------+------+
| platform|visits|
+----------+------+
| SMS| 61652|
| SMS| 44547|
| Flickr| null|
|Newsletter| null|
| Twitter| 389|
+----------+------+
only showing top 5 rows
df_save: org.apache.spark.sql.DataFrame = [platform: string, visits: int]
df: org.apache.spark.sql.DataFrame = [platform: string, visits: int]
# Loading Parquet table in Python
dfPy = spark.read.parquet("/tmp/platforms.parquet")
dfPy.show(5)
+----------+------+
| platform|visits|
+----------+------+
| SMS| 61652|
| SMS| 44547|
| Flickr| null|
|Newsletter| null|
| Twitter| 389|
+----------+------+
only showing top 5 rows
// Saving JSON dataset in Scala
val df_save = spark.table("social_media_usage").select("platform", "visits")
df_save.write.json("/tmp/platforms.json")
// Loading JSON dataset in Scala
val df = spark.read.json("/tmp/platforms.json")
df.show(5)
+----------+------+
| platform|visits|
+----------+------+
| SMS| 61652|
| SMS| 44547|
| Flickr| null|
|Newsletter| null|
| Twitter| 389|
+----------+------+
only showing top 5 rows
df_save: org.apache.spark.sql.DataFrame = [platform: string, visits: int]
df: org.apache.spark.sql.DataFrame = [platform: string, visits: bigint]
# Loading JSON dataset in Python
dfPy = spark.read.json("/tmp/platforms.json")
dfPy.show(5)
+----------+------+
| platform|visits|
+----------+------+
| SMS| 61652|
| SMS| 44547|
| Flickr| null|
|Newsletter| null|
| Twitter| 389|
+----------+------+
only showing top 5 rows
Manually Specifying Options
You can also manually specify the data source that will be used along with any extra options that you would like to pass to the data source. Data sources are specified by their fully qualified name (i.e., org.apache.spark.sql.parquet), but for built-in sources you can also use their short names (json, parquet, jdbc). DataFrames of any type can be converted into other types using this syntax.
val json = sqlContext.read.format("json").load("/tmp/platforms.json")
json.select("platform").show(10)
val parquet = sqlContext.read.format("parquet").load("/tmp/platforms.parquet")
parquet.select("platform").show(10)
+----------+
| platform|
+----------+
| SMS|
| SMS|
| Flickr|
|Newsletter|
| Twitter|
| Twitter|
| Android|
| Android|
| Android|
|Broadcastr|
+----------+
only showing top 10 rows
+----------+
| platform|
+----------+
| SMS|
| SMS|
| Flickr|
|Newsletter|
| Twitter|
| Twitter|
| Android|
| Android|
| Android|
|Broadcastr|
+----------+
only showing top 10 rows
json: org.apache.spark.sql.DataFrame = [platform: string, visits: bigint]
parquet: org.apache.spark.sql.DataFrame = [platform: string, visits: int]
Run SQL on files directly
Instead of using read API to load a file into DataFrame and query it, you can also query that file directly with SQL.
val df = sqlContext.sql("SELECT * FROM parquet.`/tmp/platforms.parquet`")
df.printSchema()
root
|-- platform: string (nullable = true)
|-- visits: integer (nullable = true)
df: org.apache.spark.sql.DataFrame = [platform: string, visits: int]
Save Modes
Save operations can optionally take a SaveMode, that specifies how to handle existing data if present. It is important to realize that these save modes do not utilize any locking and are not atomic. Additionally, when performing a Overwrite, the data will be deleted before writing out the new data.
| Scala/Java | Any language | Meaning |
|---|---|---|
SaveMode.ErrorIfExists (default) | "error" (default) | When saving a DataFrame to a data source, if data already exists, an exception is expected to be thrown. |
SaveMode.Append | "append" | When saving a DataFrame to a data source, if data/table already exists, contents of the DataFrame are expected to be appended to existing data. |
SaveMode.Overwrite | "overwrite" | Overwrite mode means that when saving a DataFrame to a data source, if data/table already exists, existing data is expected to be overwritten by the contents of the DataFrame. |
SaveMode.Ignore | "ignore" | Ignore mode means that when saving a DataFrame to a data source, if data already exists, the save operation is expected to not save the contents of the DataFrame and to not change the existing data. This is similar to a CREATE TABLE IF NOT EXISTS in SQL. |
Saving to Persistent Tables
DataFrame and Dataset can also be saved as persistent tables using the saveAsTable command. Unlike the createOrReplaceTempView command, saveAsTable will materialize the contents of the dataframe and create a pointer to the data in the metastore. Persistent tables will still exist even after your Spark program has restarted, as long as you maintain your connection to the same metastore. A DataFrame for a persistent table can be created by calling the table method on a SparkSession with the name of the table.
By default saveAsTable will create a “managed table”, meaning that the location of the data will be controlled by the metastore. Managed tables will also have their data deleted automatically when a table is dropped.
// First of all list tables to see that table we are about to create does not exist
spark.catalog.listTables.show()
+--------------------+--------+-----------+---------+-----------+
| name|database|description|tableType|isTemporary|
+--------------------+--------+-----------+---------+-----------+
| cities_csv| default| null| EXTERNAL| false|
| cleaned_taxes| default| null| MANAGED| false|
|commdettrumpclint...| default| null| MANAGED| false|
| donaldtrumptweets| default| null| EXTERNAL| false|
| linkage| default| null| EXTERNAL| false|
| nations| default| null| EXTERNAL| false|
| newmplist| default| null| EXTERNAL| false|
| ny_baby_names| default| null| MANAGED| false|
| nzmpsandparty| default| null| EXTERNAL| false|
| pos_neg_category| default| null| EXTERNAL| false|
| rna| default| null| MANAGED| false|
| samh| default| null| EXTERNAL| false|
| social_media_usage| default| null| EXTERNAL| false|
| table1| default| null| EXTERNAL| false|
| test_table| default| null| EXTERNAL| false|
| uscites| default| null| EXTERNAL| false|
+--------------------+--------+-----------+---------+-----------+
drop table if exists simple_range
val df = spark.range(0, 100)
df.write.saveAsTable("simple_range")
// Verify that table is saved and it is marked as persistent ("isTemporary" value should be "false")
spark.catalog.listTables.show()
+--------------------+--------+-----------+---------+-----------+
| name|database|description|tableType|isTemporary|
+--------------------+--------+-----------+---------+-----------+
| cities_csv| default| null| EXTERNAL| false|
| cleaned_taxes| default| null| MANAGED| false|
|commdettrumpclint...| default| null| MANAGED| false|
| donaldtrumptweets| default| null| EXTERNAL| false|
| linkage| default| null| EXTERNAL| false|
| nations| default| null| EXTERNAL| false|
| newmplist| default| null| EXTERNAL| false|
| ny_baby_names| default| null| MANAGED| false|
| nzmpsandparty| default| null| EXTERNAL| false|
| pos_neg_category| default| null| EXTERNAL| false|
| rna| default| null| MANAGED| false|
| samh| default| null| EXTERNAL| false|
| simple_range| default| null| MANAGED| false|
| social_media_usage| default| null| EXTERNAL| false|
| table1| default| null| EXTERNAL| false|
| test_table| default| null| EXTERNAL| false|
| uscites| default| null| EXTERNAL| false|
+--------------------+--------+-----------+---------+-----------+
df: org.apache.spark.sql.Dataset[Long] = [id: bigint]
Parquet Files
Parquet is a columnar format that is supported by many other data processing systems. Spark SQL provides support for both reading and writing Parquet files that automatically preserves the schema of the original data. When writing Parquet files, all columns are automatically converted to be nullable for compatibility reasons.
More on Parquet
Apache Parquet is a columnar storage format available to any project in the Hadoop ecosystem, regardless of the choice of data processing framework, data model or programming language. It is a more efficient way to store data frames.
- To understand the ideas read Dremel: Interactive Analysis of Web-Scale Datasets, Sergey Melnik, Andrey Gubarev, Jing Jing Long, Geoffrey Romer, Shiva Shivakumar, Matt Tolton and Theo Vassilakis,Proc. of the 36th Int'l Conf on Very Large Data Bases (2010), pp. 330-339, whose Abstract is as follows:
- Dremel is a scalable, interactive ad-hoc query system for analysis of read-only nested data. By combining multi-level execution trees and columnar data layouts it is capable of running aggregation queries over trillion-row tables in seconds. The system scales to thousands of CPUs and petabytes of data, and has thousands of users at Google. In this paper, we describe the architecture and implementation of Dremel, and explain how it complements MapReduce-based computing. We present a novel columnar storage representation for nested records and discuss experiments on few-thousand node instances of the system.
// Read in the parquet file created above. Parquet files are self-describing so the schema is preserved.
// The result of loading a Parquet file is also a DataFrame.
val parquetFile = sqlContext.read.parquet("/tmp/platforms.parquet")
// Parquet files can also be registered as tables and then used in SQL statements.
parquetFile.createOrReplaceTempView("parquetFile")
val platforms = sqlContext.sql("SELECT platform FROM parquetFile WHERE visits > 0")
platforms.distinct.map(t => "Name: " + t(0)).collect().foreach(println)
Name: Flickr
Name: iPhone
Name: YouTube
Name: WordPress
Name: SMS
Name: iPhone App
Name: Youtube
Name: Instagram
Name: iPhone app
Name: Linked-In
Name: Twitter
Name: TOTAL
Name: Tumblr
Name: Newsletter
Name: Pinterest
Name: Android
Name: Foursquare
Name: Google+
Name: Foursquare (Badge Unlock)
Name: Facebook
parquetFile: org.apache.spark.sql.DataFrame = [platform: string, visits: int]
platforms: org.apache.spark.sql.DataFrame = [platform: string]
Partition Discovery
Table partitioning is a common optimization approach used in systems like Hive. In a partitioned table, data are usually stored in different directories, with partitioning column values encoded in the path of each partition directory. The Parquet data source is now able to discover and infer partitioning information automatically. For example, we can store all our previously used population data (from the programming guide example!) into a partitioned table using the following directory structure, with two extra columns, gender and country as partitioning columns:
path
└── to
└── table
├── gender=male
│ ├── ...
│ │
│ ├── country=US
│ │ └── data.parquet
│ ├── country=CN
│ │ └── data.parquet
│ └── ...
└── gender=female
├── ...
│
├── country=US
│ └── data.parquet
├── country=CN
│ └── data.parquet
└── ...
By passing path/to/table to either SparkSession.read.parquet or SparkSession.read.load, Spark SQL will automatically extract the partitioning information from the paths. Now the schema of the returned DataFrame becomes:
root
|-- name: string (nullable = true)
|-- age: long (nullable = true)
|-- gender: string (nullable = true)
|-- country: string (nullable = true)
Notice that the data types of the partitioning columns are automatically inferred. Currently, numeric data types and string type are supported. Sometimes users may not want to automatically infer the data types of the partitioning columns. For these use cases, the automatic type inference can be configured by spark.sql.sources.partitionColumnTypeInference.enabled, which is default to true. When type inference is disabled, string type will be used for the partitioning columns.
Starting from Spark 1.6.0, partition discovery only finds partitions under the given paths by default. For the above example, if users pass path/to/table/gender=male to either SparkSession.read.parquet or SparkSession.read.load, gender will not be considered as a partitioning column. If users need to specify the base path that partition discovery should start with, they can set basePath in the data source options. For example, when path/to/table/gender=male is the path of the data and users set basePath to path/to/table/, gender will be a partitioning column.
Schema Merging
Like ProtocolBuffer, Avro, and Thrift, Parquet also supports schema evolution. Users can start with a simple schema, and gradually add more columns to the schema as needed. In this way, users may end up with multiple Parquet files with different but mutually compatible schemas. The Parquet data source is now able to automatically detect this case and merge schemas of all these files.
Since schema merging is a relatively expensive operation, and is not a necessity in most cases, we turned it off by default starting from 1.5.0. You may enable it by: 1. setting data source option mergeSchema to true when reading Parquet files (as shown in the examples below), or 2. setting the global SQL option spark.sql.parquet.mergeSchema to true.
// Create a simple DataFrame, stored into a partition directory
val df1 = sc.parallelize(1 to 5).map(i => (i, i * 2)).toDF("single", "double")
df1.write.mode("overwrite").parquet("/tmp/data/test_table/key=1")
// Create another DataFrame in a new partition directory, adding a new column and dropping an existing column
val df2 = sc.parallelize(6 to 10).map(i => (i, i * 3)).toDF("single", "triple")
df2.write.mode("overwrite").parquet("/tmp/data/test_table/key=2")
// Read the partitioned table
val df3 = spark.read.option("mergeSchema", "true").parquet("/tmp/data/test_table")
df3.printSchema()
// The final schema consists of all 3 columns in the Parquet files together
// with the partitioning column appeared in the partition directory paths.
// root
// |-- single: integer (nullable = true)
// |-- double: integer (nullable = true)
// |-- triple: integer (nullable = true)
// |-- key: integer (nullable = true))
root
|-- single: integer (nullable = true)
|-- double: integer (nullable = true)
|-- triple: integer (nullable = true)
|-- key: integer (nullable = true)
df1: org.apache.spark.sql.DataFrame = [single: int, double: int]
df2: org.apache.spark.sql.DataFrame = [single: int, triple: int]
df3: org.apache.spark.sql.DataFrame = [single: int, double: int ... 2 more fields]
Hive metastore Parquet table conversion
When reading from and writing to Hive metastore Parquet tables, Spark SQL will try to use its own Parquet support instead of Hive SerDe for better performance. This behavior is controlled by the spark.sql.hive.convertMetastoreParquet configuration, and is turned on by default.
Hive/Parquet Schema Reconciliation
There are two key differences between Hive and Parquet from the perspective of table schema processing. 1. Hive is case insensitive, while Parquet is not 2. Hive considers all columns nullable, while nullability in Parquet is significant
Due to this reason, we must reconcile Hive metastore schema with Parquet schema when converting a Hive metastore Parquet table to a Spark SQL Parquet table. The reconciliation rules are: 1. Fields that have the same name in both schema must have the same data type regardless of nullability. The reconciled field should have the data type of the Parquet side, so that nullability is respected. 2. The reconciled schema contains exactly those fields defined in Hive metastore schema. - Any fields that only appear in the Parquet schema are dropped in the reconciled schema. - Any fileds that only appear in the Hive metastore schema are added as nullable field in the reconciled schema.
Metadata Refreshing
Spark SQL caches Parquet metadata for better performance. When Hive metastore Parquet table conversion is enabled, metadata of those converted tables are also cached. If these tables are updated by Hive or other external tools, you need to refresh them manually to ensure consistent metadata.
// should refresh table metadata
spark.catalog.refreshTable("simple_range")
-- Or you can use SQL to refresh table
REFRESH TABLE simple_range;
Configuration
Configuration of Parquet can be done using the setConf method on SQLContext or by running SET key=value commands using SQL.
| Property Name | Default | Meaning | |
|---|---|---|---|
spark.sql.parquet.binaryAsString | false | Some other Parquet-producing systems, in particular Impala, Hive, and older versions of Spark SQL, do not differentiate between binary data and strings when writing out the Parquet schema. This flag tells Spark SQL to interpret binary data as a string to provide compatibility with these systems. | |
spark.sql.parquet.int96AsTimestamp | true | Some Parquet-producing systems, in particular Impala and Hive, store Timestamp into INT96. This flag tells Spark SQL to interpret INT96 data as a timestamp to provide compatibility with these systems. | |
spark.sql.parquet.cacheMetadata | true | Turns on caching of Parquet schema metadata. Can speed up querying of static data. | |
spark.sql.parquet.compression.codec | gzip | Sets the compression codec use when writing Parquet files. Acceptable values include: uncompressed, snappy, gzip, lzo. | |
spark.sql.parquet.filterPushdown | true | Enables Parquet filter push-down optimization when set to true. | |
spark.sql.hive.convertMetastoreParquet | true | When set to false, Spark SQL will use the Hive SerDe for parquet tables instead of the built in support. | |
spark.sql.parquet.output.committer.class | org.apache.parquet.hadoop.ParquetOutputCommitter | The output committer class used by Parquet. The specified class needs to be a subclass of org.apache.hadoop.mapreduce.OutputCommitter. Typically, it's also a subclass of org.apache.parquet.hadoop.ParquetOutputCommitter. Spark SQL comes with a builtin org.apache.spark.sql.parquet.DirectParquetOutputCommitter, which can be more efficient then the default Parquet output committer when writing data to S3. | |
spark.sql.parquet.mergeSchema | false | When true, the Parquet data source merges schemas collected from all data files, otherwise the schema is picked from the summary file or a random data file if no summary file is available. |
JSON Datasets
Spark SQL can automatically infer the schema of a JSON dataset and load it as a DataFrame. This conversion can be done using SparkSession.read.json() on either an RDD of String, or a JSON file.
Note that the file that is offered as a json file is not a typical JSON file. Each line must contain a separate, self-contained valid JSON object. As a consequence, a regular multi-line JSON file will most often fail.
// A JSON dataset is pointed to by path.
// The path can be either a single text file or a directory storing text files.
val path = "/tmp/platforms.json"
val platforms = spark.read.json(path)
// The inferred schema can be visualized using the printSchema() method.
platforms.printSchema()
// root
// |-- platform: string (nullable = true)
// |-- visits: long (nullable = true)
// Register this DataFrame as a table.
platforms.createOrReplaceTempView("platforms")
// SQL statements can be run by using the sql methods provided by sqlContext.
val facebook = spark.sql("SELECT platform, visits FROM platforms WHERE platform like 'Face%k'")
facebook.show()
// Alternatively, a DataFrame can be created for a JSON dataset represented by
// an RDD[String] storing one JSON object per string.
val rdd = sc.parallelize("""{"name":"IWyn","address":{"city":"Columbus","state":"Ohio"}}""" :: Nil)
val anotherPlatforms = spark.read.json(rdd)
anotherPlatforms.show()
root
|-- platform: string (nullable = true)
|-- visits: long (nullable = true)
+--------+------+
|platform|visits|
+--------+------+
|Facebook| 3|
|Facebook| 36|
|Facebook| 47|
|Facebook| 90|
|Facebook| 105|
|Facebook| 123|
|Facebook| 119|
|Facebook| 148|
|Facebook| 197|
|Facebook| 179|
|Facebook| 200|
|Facebook| 200|
|Facebook| 183|
|Facebook| 190|
|Facebook| 227|
|Facebook| 194|
|Facebook| 243|
|Facebook| 237|
|Facebook| 234|
|Facebook| 238|
+--------+------+
only showing top 20 rows
+---------------+----+
| address|name|
+---------------+----+
|[Columbus,Ohio]|IWyn|
+---------------+----+
<console>:63: warning: method json in class DataFrameReader is deprecated: Use json(Dataset[String]) instead.
val anotherPlatforms = spark.read.json(rdd)
^
path: String = /tmp/platforms.json
platforms: org.apache.spark.sql.DataFrame = [platform: string, visits: bigint]
facebook: org.apache.spark.sql.DataFrame = [platform: string, visits: bigint]
rdd: org.apache.spark.rdd.RDD[String] = ParallelCollectionRDD[23304] at parallelize at <console>:62
anotherPlatforms: org.apache.spark.sql.DataFrame = [address: struct<city: string, state: string>, name: string]
Hive Tables
Spark SQL also supports reading and writing data stored in Apache Hive. However, since Hive has a large number of dependencies, it is not included in the default Spark assembly. Hive support is enabled by adding the -Phive and -Phive-thriftserver flags to Spark’s build. This command builds a new assembly jar that includes Hive. Note that this Hive assembly jar must also be present on all of the worker nodes, as they will need access to the Hive serialization and deserialization libraries (SerDes) in order to access data stored in Hive.
Configuration of Hive is done by placing your hive-site.xml, core-site.xml (for security configuration), hdfs-site.xml (for HDFS configuration) file in conf/. Please note when running the query on a YARN cluster (cluster mode), the datanucleus jars under the lib_managed/jars directory and hive-site.xml under conf/ directory need to be available on the driver and all executors launched by the YARN cluster. The convenient way to do this is adding them through the --jars option and --file option of the spark-submit command.
When working with Hive one must construct a HiveContext, which inherits from SQLContext, and adds support for finding tables in the MetaStore and writing queries using HiveQL. Users who do not have an existing Hive deployment can still create a HiveContext. When not configured by the hive-site.xml, the context automatically creates metastore_db in the current directory and creates warehouse directory indicated by HiveConf, which defaults to /user/hive/warehouse. Note that you may need to grant write privilege on /user/hive/warehouse to the user who starts the spark application.
val spark = SparkSession.builder.enableHiveSupport().getOrCreate()
spark.sql("CREATE TABLE IF NOT EXISTS src (key INT, value STRING)")
spark.sql("LOAD DATA LOCAL INPATH 'examples/src/main/resources/kv1.txt' INTO TABLE src")
// Queries are expressed in HiveQL
spark.sql("FROM src SELECT key, value").collect().foreach(println)
Interacting with Different Versions of Hive Metastore
One of the most important pieces of Spark SQL’s Hive support is interaction with Hive metastore, which enables Spark SQL to access metadata of Hive tables. Starting from Spark 1.4.0, a single binary build of Spark SQL can be used to query different versions of Hive metastores, using the configuration described below. Note that independent of the version of Hive that is being used to talk to the metastore, internally Spark SQL will compile against Hive 1.2.1 and use those classes for internal execution (serdes, UDFs, UDAFs, etc).
The following options can be used to configure the version of Hive that is used to retrieve metadata:
| Property Name | Default | Meaning |
|---|---|---|
spark.sql.hive.metastore.version | 1.2.1 | Version of the Hive metastore. Available options are 0.12.0 through 1.2.1. |
spark.sql.hive.metastore.jars | builtin | Location of the jars that should be used to instantiate the HiveMetastoreClient. This property can be one of three options: builtin, maven, a classpath in the standard format for the JVM. This classpath must include all of Hive and its dependencies, including the correct version of Hadoop. These jars only need to be present on the driver, but if you are running in yarn cluster mode then you must ensure they are packaged with you application. |
spark.sql.hive.metastore.sharedPrefixes | com.mysql.jdbc,org.postgresql,com.microsoft.sqlserver,oracle.jdbc | A comma separated list of class prefixes that should be loaded using the classloader that is shared between Spark SQL and a specific version of Hive. An example of classes that should be shared is JDBC drivers that are needed to talk to the metastore. Other classes that need to be shared are those that interact with classes that are already shared. For example, custom appenders that are used by log4j. |
spark.sql.hive.metastore.barrierPrefixes | (empty) | A comma separated list of class prefixes that should explicitly be reloaded for each version of Hive that Spark SQL is communicating with. For example, Hive UDFs that are declared in a prefix that typically would be shared (i.e. org.apache.spark.*). |
JDBC To Other Databases
Spark SQL also includes a data source that can read data from other databases using JDBC. This functionality should be preferred over using JdbcRDD. This is because the results are returned as a DataFrame and they can easily be processed in Spark SQL or joined with other data sources. The JDBC data source is also easier to use from Java or Python as it does not require the user to provide a ClassTag. (Note that this is different than the Spark SQL JDBC server, which allows other applications to run queries using Spark SQL).
To get started you will need to include the JDBC driver for you particular database on the spark classpath. For example, to connect to postgres from the Spark Shell you would run the following command:
SPARK_CLASSPATH=postgresql-9.3-1102-jdbc41.jar bin/spark-shell
Tables from the remote database can be loaded as a DataFrame or Spark SQL Temporary table using the Data Sources API. The following options are supported:
| Property Name | Meaning | |
|---|---|---|
url | The JDBC URL to connect to. | |
dbtable | The JDBC table that should be read. Note that anything that is valid in a FROM clause of a SQL query can be used. For example, instead of a full table you could also use a subquery in parentheses. | |
driver | The class name of the JDBC driver needed to connect to this URL. This class will be loaded on the master and workers before running an JDBC commands to allow the driver to register itself with the JDBC subsystem. | |
partitionColumn, lowerBound, upperBound, numPartitions | These options must all be specified if any of them is specified. They describe how to partition the table when reading in parallel from multiple workers. partitionColumn must be a numeric column from the table in question. Notice that lowerBound and upperBound are just used to decide the partition stride, not for filtering the rows in table. So all rows in the table will be partitioned and returned. | |
fetchSize | The JDBC fetch size, which determines how many rows to fetch per round trip. This can help performance on JDBC drivers which default to low fetch size (eg. Oracle with 10 rows). |
// Example of using JDBC datasource
val jdbcDF = spark.read.format("jdbc").options(Map("url" -> "jdbc:postgresql:dbserver", "dbtable" -> "schema.tablename")).load()
-- Or using JDBC datasource in SQL
CREATE TEMPORARY TABLE jdbcTable
USING org.apache.spark.sql.jdbc
OPTIONS (
url "jdbc:postgresql:dbserver",
dbtable "schema.tablename"
)
Troubleshooting
- The JDBC driver class must be visible to the primordial class loader on the client session and on all executors. This is because Java’s DriverManager class does a security check that results in it ignoring all drivers not visible to the primordial class loader when one goes to open a connection. One convenient way to do this is to modify compute_classpath.sh on all worker nodes to include your driver JARs.
- Some databases, such as H2, convert all names to upper case. You’ll need to use upper case to refer to those names in Spark SQL.
This is an elaboration of the Apache Spark 2.2 sql-progamming-guide.
Performance Tuning
Spark Sql Programming Guide
- Overview
- SQL
- DataFrames
- Datasets
- Getting Started
- Starting Point: SQLContext
- Creating DataFrames
- DataFrame Operations
- Running SQL Queries Programmatically
- Creating Datasets
- Interoperating with RDDs
- Inferring the Schema Using Reflection
- Programmatically Specifying the Schema
- Data Sources
- Generic Load/Save Functions
- Manually Specifying Options
- Run SQL on files directly
- Save Modes
- Saving to Persistent Tables
- Parquet Files
- Loading Data Programmatically
- Partition Discovery
- Schema Merging
- Hive metastore Parquet table conversion
- Hive/Parquet Schema Reconciliation
- Metadata Refreshing
- Configuration
- JSON Datasets
- Hive Tables
- Interacting with Different Versions of Hive Metastore
- JDBC To Other Databases
- Troubleshooting
- Generic Load/Save Functions
- Performance Tuning
- Caching Data In Memory
- Other Configuration Options
- Distributed SQL Engine
- Running the Thrift JDBC/ODBC server
- Running the Spark SQL CLI
Performance Tuning
For some workloads it is possible to improve performance by either caching data in memory, or by turning on some experimental options.
Caching Data In Memory
Spark SQL can cache tables using an in-memory columnar format by calling spark.cacheTable("tableName") or dataset.cache(). Then Spark SQL will scan only required columns and will automatically tune compression to minimize memory usage and GC pressure. You can call spark.uncacheTable("tableName") to remove the table from memory.
Configuration of in-memory caching can be done using the setConf method on SparkSession or by running SET key=value commands using SQL.
| Property Name | Default | Meaning |
|---|---|---|
spark.sql.inMemoryColumnarStorage.compressed | true | When set to true Spark SQL will automatically select a compression codec for each column based on statistics of the data. |
spark.sql.inMemoryColumnarStorage.batchSize | 10000 | Controls the size of batches for columnar caching. Larger batch sizes can improve memory utilization and compression, but risk OOMs when caching data. |
Other Configuration Options
The following options can also be used to tune the performance of query execution. It is possible that these options will be deprecated in future release as more optimizations are performed automatically.
| Property Name | Default | Meaning |
|---|---|---|
spark.sql.autoBroadcastJoinThreshold | 10485760 (10 MB) | Configures the maximum size in bytes for a table that will be broadcast to all worker nodes when performing a join. By setting this value to -1 broadcasting can be disabled. Note that currently statistics are only supported for Hive Metastore tables where the command ANALYZE TABLE <tableName> COMPUTE STATISTICS noscan has been run. |
spark.sql.tungsten.enabled | true | When true, use the optimized Tungsten physical execution backend which explicitly manages memory and dynamically generates bytecode for expression evaluation. |
spark.sql.shuffle.partitions | 200 | Configures the number of partitions to use when shuffling data for joins or aggregations. |
This is an elaboration of the Apache Spark 2.2 sql-progamming-guide.
Distributed SQL Engine
Spark Sql Programming Guide
- Overview
- SQL
- DataFrames
- Datasets
- Getting Started
- Starting Point: SQLContext
- Creating DataFrames
- DataFrame Operations
- Running SQL Queries Programmatically
- Creating Datasets
- Interoperating with RDDs
- Inferring the Schema Using Reflection
- Programmatically Specifying the Schema
- Data Sources
- Generic Load/Save Functions
- Manually Specifying Options
- Run SQL on files directly
- Save Modes
- Saving to Persistent Tables
- Parquet Files
- Loading Data Programmatically
- Partition Discovery
- Schema Merging
- Hive metastore Parquet table conversion
- Hive/Parquet Schema Reconciliation
- Metadata Refreshing
- Configuration
- JSON Datasets
- Hive Tables
- Interacting with Different Versions of Hive Metastore
- JDBC To Other Databases
- Troubleshooting
- Generic Load/Save Functions
- Performance Tuning
- Caching Data In Memory
- Other Configuration Options
- Distributed SQL Engine
- Running the Thrift JDBC/ODBC server
- Running the Spark SQL CLI
Distributed SQL Engine
Spark SQL can also act as a distributed query engine using its JDBC/ODBC or command-line interface. In this mode, end-users or applications can interact with Spark SQL directly to run SQL queries, without the need to write any code.
Running the Thrift JDBC/ODBC server
The Thrift JDBC/ODBC server implemented here corresponds to the HiveServer2 in Hive 1.2.1 You can test the JDBC server with the beeline script that comes with either Spark or Hive 1.2.1.
To start the JDBC/ODBC server, run the following in the Spark directory:
./sbin/start-thriftserver.sh
This script accepts all bin/spark-submit command line options, plus a --hiveconf option to specify Hive properties. You may run ./sbin/start-thriftserver.sh --help for a complete list of all available options. By default, the server listens on localhost:10000. You may override this behaviour via either environment variables, i.e.:
export HIVE_SERVER2_THRIFT_PORT=<listening-port>
export HIVE_SERVER2_THRIFT_BIND_HOST=<listening-host>
./sbin/start-thriftserver.sh \
--master <master-uri> \
...
or system properties:
./sbin/start-thriftserver.sh \
--hiveconf hive.server2.thrift.port=<listening-port> \
--hiveconf hive.server2.thrift.bind.host=<listening-host> \
--master <master-uri>
...
Now you can use beeline to test the Thrift JDBC/ODBC server:
./bin/beeline
Connect to the JDBC/ODBC server in beeline with:
beeline> !connect jdbc:hive2://localhost:10000
Beeline will ask you for a username and password. In non-secure mode, simply enter the username on your machine and a blank password. For secure mode, please follow the instructions given in the beeline documentation.
Configuration of Hive is done by placing your hive-site.xml, core-site.xml and hdfs-site.xml files in conf/.
You may also use the beeline script that comes with Hive.
Thrift JDBC server also supports sending thrift RPC messages over HTTP transport. Use the following setting to enable HTTP mode as system property or in hive-site.xml file in conf/:
hive.server2.transport.mode - Set this to value: http
hive.server2.thrift.http.port - HTTP port number fo listen on; default is 10001
hive.server2.http.endpoint - HTTP endpoint; default is cliservice
To test, use beeline to connect to the JDBC/ODBC server in http mode with:
beeline> !connect jdbc:hive2://<host>:<port>/<database>?hive.server2.transport.mode=http;hive.server2.thrift.http.path=<http_endpoint>
Running the Spark SQL CLI
The Spark SQL CLI is a convenient tool to run the Hive metastore service in local mode and execute queries input from the command line. Note that the Spark SQL CLI cannot talk to the Thrift JDBC server.
To start the Spark SQL CLI, run the following in the Spark directory:
./bin/spark-sql
Configuration of Hive is done by placing your hive-site.xml, core-site.xml and hdfs-site.xml files in conf/. You may run ./bin/spark-sql --help for a complete list of all available options.
Extract, Transform and Load (ETL) of the SoU Addresses
A bit of bash and lynx to achieve the scraping of the state of the union addresses of the US Presidents
by Paul Brouwers
And some Shell-level parsed-data exploration, injection into the distributed file system and testing
by Raazesh Sainudiin
This SoU dataset is used in the 006_WordCount notebook.
The code below is mainly there to show how the text content of each state of the union address was scraped from the following URL: * http://stateoftheunion.onetwothree.net/texts/index.html
Such data acquisition task or ETL is usually the first and crucial step in a data scientist's workflow. A data scientist generally does the scraping and parsing of the data by her/himself. Data ingestion not only allows the scientist to start the analysis but also determines the quality of the analysis by the limits it imposes on the accessible feature space.
We have done this and put the data in the distributed file system for easy loading into our notebooks for further analysis. This keeps us from having to install unix programs like lynx, sed, etc. that are needed in the shell script below.
for i in $(lynx --dump http://stateoftheunion.onetwothree.net/texts/index.html | grep texts | grep -v index | sed 's/.*http/http/') ; do lynx --dump $i | tail -n+13 | head -n-14 | sed 's/^\s\+//' | sed -e ':a;N;$!ba;s/\(.\)\n/\1 /g' -e 's/\n/\n\n/' > $(echo $i | sed 's/.*\([0-9]\{8\}\).*/\1/').txt ; done
Or in a more atomic form:
for i in $(lynx --dump http://stateoftheunion.onetwothree.net/texts/index.html \
| grep texts \
| grep -v index \
| sed 's/.*http/http/')
do
lynx --dump $i \
| tail -n+13 \
| head -n-14 \
| sed 's/^\s\+//' \
| sed -e ':a;N;$!ba;s/\(.\)\n/\1 /g' -e 's/\n/\n\n/' \
> $(echo $i | sed 's/.*\([0-9]\{8\}\).*/\1/').txt
done
Don't re-evaluate!
The following BASH (shell) script can be made to work on databricks cloud directly by installing the dependencies such as lynx, etc. Since we have already scraped it and put the data in our distributed file system let's not evaluate or <Ctrl+Enter> the cell below. The cell is mainly there to show how it can be done (you may want to modify it to scrape other sites for other text data).
#remove the hash character from the line below to evaluate when needed
#for i in $(lynx --dump http://stateoftheunion.onetwothree.net/texts/index.html | grep texts | grep -v index | sed 's/.*http/http/') ; do lynx --dump $i | tail -n+13 | head -n-14 | sed 's/^\s\+//' | sed -e ':a;N;$!ba;s/\(.\)\n/\1 /g' -e 's/\n/\n\n/' > $(echo $i | sed 's/.*\([0-9]\{8\}\).*/\1/').txt ; done
pwd && ls && du -sh .
/databricks/driver
conf
derby.log
eventlogs
ganglia
logs
5.2M .
ls /home/ubuntu && du -sh /home/ubuntu
databricks
40K /home/ubuntu
We can just grab the data as a tarball (gnuZipped tar archive) file sou.tar.gz using wget as follows:
%sh wget http://www.math.canterbury.ac.nz/~r.sainudiin/datasets/public/SOU/sou.tar.gz
df -h
pwd
Filesystem Size Used Avail Use% Mounted on
/var/lib/lxc/base-images/release__7.2.x-snapshot-scala2.12__databricks-universe__head__0652b44__47ac4a2__jenkins__0b22b70__format-2 148G 11G 130G 8% /
none 492K 0 492K 0% /dev
udev 3.9G 0 3.9G 0% /dev/tty
/dev/xvdb 148G 11G 130G 8% /mnt/readonly
/dev/mapper/vg-lv 296G 11G 271G 4% /local_disk0
tmpfs 3.9G 0 3.9G 0% /dev/shm
tmpfs 3.9G 232K 3.9G 1% /run
tmpfs 5.0M 0 5.0M 0% /run/lock
tmpfs 3.9G 0 3.9G 0% /sys/fs/cgroup
tmpfs 315M 0 315M 0% /run/user/1000
/: 1.0P 0 1.0P 0% /dbfs
/databricks/driver
pwd
ls
/databricks/driver
conf
derby.log
eventlogs
ganglia
logs
wget http://lamastex.org/datasets/public/SOU/README.md
cat README.md
--2020-09-16 18:49:05-- http://lamastex.org/datasets/public/SOU/README.md
Resolving lamastex.org (lamastex.org)... 166.62.28.100
Connecting to lamastex.org (lamastex.org)|166.62.28.100|:80... connected.
HTTP request sent, awaiting response... 200 OK
Length: 8087 (7.9K)
Saving to: ‘README.md’
0K ....... 100% 553M=0s
2020-09-16 18:49:06 (553 MB/s) - ‘README.md’ saved [8087/8087]
## A bit of bash and lynx to achieve the scraping of the state of the union addresses of the US Presidents
### by Paul Brouwers
The code below is mainly there to show how the text content of each state of the union address was scraped from the following URL:
* [http://stateoftheunion.onetwothree.net/texts/index.html](http://stateoftheunion.onetwothree.net/texts/index.html)
Such data acquisition tasks is usually the first and cucial step in a data scientist's workflow.
We have done this and put the data in the distributed file system for easy loading into our notebooks for further analysis. This keeps us from having to install unix programs like ``lynx``, ``sed``, etc. that are needed in the shell script below.
```%sh
for i in $(lynx --dump http://stateoftheunion.onetwothree.net/texts/index.html | grep texts | grep -v index | sed 's/.*http/http/') ; do lynx --dump $i | tail -n+13 | head -n-14 | sed 's/^\s\+//' | sed -e ':a;N;$!ba;s/\(.\)\n/\1 /g' -e 's/\n/\n\n/' > $(echo $i | sed 's/.*\([0-9]\{8\}\).*/\1/').txt ; done
```
Or in a more atomic form:
```%sh
for i in $(lynx --dump http://stateoftheunion.onetwothree.net/texts/index.html \
| grep texts \
| grep -v index \
| sed 's/.*http/http/')
do
lynx --dump $i \
| tail -n+13 \
| head -n-14 \
| sed 's/^\s\+//' \
| sed -e ':a;N;$!ba;s/\(.\)\n/\1 /g' -e 's/\n/\n\n/' \
> $(echo $i | sed 's/.*\([0-9]\{8\}\).*/\1/').txt
done
```
**Don't re-evaluate!**
The following BASH (shell) script can be made to work on databricks cloud directly by installing the dependencies such as ``lynx``, etc. Since we have already scraped it and put the data in our distributed file system **let's not evaluate or ``<Ctrl+Enter>`` the cell below**. The cell is mainly there to show how it can be done (you may want to modify it to scrape other sites for other text data).
```%sh
#remove the hash character from the line below to evaluate when needed
#for i in $(lynx --dump http://stateoftheunion.onetwothree.net/texts/index.html | grep texts | grep -v index | sed 's/.*http/http/') ; do lynx --dump $i | tail -n+13 | head -n-14 | sed 's/^\s\+//' | sed -e ':a;N;$!ba;s/\(.\)\n/\1 /g' -e 's/\n/\n\n/' > $(echo $i | sed 's/.*\([0-9]\{8\}\).*/\1/').txt ; done
```
We can just grab the data as a tarball (gnuZipped tar archive) file ``sou.tar.gz`` using wget as follows:
```%sh
wget http://www.math.canterbury.ac.nz/~r.sainudiin/datasets/public/SOU/sou.tar.gz
```
Raazesh Sainudiin
Fri Feb 19 11:08:00 NZDT 2016
# Atlantis Redo
Redone to include President Trump's SOU address in 2017.
```%sh
zesh@raazesh-Inspiron-15-7579:~$ sudo apt-get -y install lynx
[sudo] password for raazesh:
Reading package lists... Done
Building dependency tree
Reading state information... Done
The following additional packages will be installed:
lynx-common
The following NEW packages will be installed:
lynx lynx-common
0 upgraded, 2 newly installed, 0 to remove and 0 not upgraded.
Need to get 1,035 kB of archives.
After this operation, 2,761 kB of additional disk space will be used.
Get:1 http://ubuntu.mirror.su.se/ubuntu xenial/universe amd64 lynx-common all 2.8.9dev8-4ubuntu1 [411 kB]
Get:2 http://ubuntu.mirror.su.se/ubuntu xenial/universe amd64 lynx amd64 2.8.9dev8-4ubuntu1 [624 kB]
Fetched 1,035 kB in 0s (9,643 kB/s)
Selecting previously unselected package lynx-common.
(Reading database ... 459514 files and directories currently installed.)
Preparing to unpack .../lynx-common_2.8.9dev8-4ubuntu1_all.deb ...
Unpacking lynx-common (2.8.9dev8-4ubuntu1) ...
Selecting previously unselected package lynx.
Preparing to unpack .../lynx_2.8.9dev8-4ubuntu1_amd64.deb ...
Unpacking lynx (2.8.9dev8-4ubuntu1) ...
Processing triggers for mime-support (3.59ubuntu1) ...
Processing triggers for doc-base (0.10.7) ...
Processing 1 added doc-base file...
Processing triggers for man-db (2.7.5-1) ...
Setting up lynx-common (2.8.9dev8-4ubuntu1) ...
Setting up lynx (2.8.9dev8-4ubuntu1) ...
update-alternatives: using /usr/bin/lynx to provide /usr/bin/www-browser (www-browser) in auto mode
raazesh@raazesh-Inspiron-15-7579:~$ mkdir sou
raazesh@raazesh-Inspiron-15-7579:~$ cd sou/
raazesh@raazesh-Inspiron-15-7579:~/sou$ for i in $(lynx --dump http://stateoftheunion.onetwothree.net/texts/index.html | grep texts | grep -v index | sed 's/.*http/http/') ; do lynx --dump $i | tail -n+13 | head -n-14 | sed 's/^\s\+//' | sed -e ':a;N;$!ba;s/\(.\)\n/\1 /g' -e 's/\n/\n\n/' > $(echo $i | sed 's/.*\([0-9]\{8\}\).*/\1/').txt ; done
raazesh@raazesh-Inspiron-15-7579:~/sou$ ls
17900108.txt 18101205.txt 18311206.txt 18521206.txt 18731201.txt 18941202.txt 19151207.txt 19370106.txt 19570110.txt 19770112.txt 19980127.txt
17901208.txt 18111105.txt 18321204.txt 18531205.txt 18741207.txt 18951207.txt 19161205.txt 19380103.txt 19580109.txt 19780119.txt 19990119.txt
17911025.txt 18121104.txt 18331203.txt 18541204.txt 18751207.txt 18961204.txt 19171204.txt 19390104.txt 19590109.txt 19790125.txt 20000127.txt
17921106.txt 18131207.txt 18341201.txt 18551231.txt 18761205.txt 18971206.txt 19181202.txt 19400103.txt 19600107.txt 19800121.txt 20010227.txt
17931203.txt 18140920.txt 18351207.txt 18561202.txt 18771203.txt 18981205.txt 19191202.txt 19410106.txt 19610112.txt 19810116.txt 20010920.txt
17941119.txt 18151205.txt 18361205.txt 18571208.txt 18781202.txt 18991205.txt 19201207.txt 19420106.txt 19610130.txt 19820126.txt 20020129.txt
17951208.txt 18161203.txt 18371205.txt 18581206.txt 18791201.txt 19001203.txt 19211206.txt 19430107.txt 19620111.txt 19830125.txt 20030128.txt
17961207.txt 18171212.txt 18381203.txt 18591219.txt 18801206.txt 19011203.txt 19221208.txt 19440111.txt 19630114.txt 19840125.txt 20040120.txt
17971122.txt 18181116.txt 18391202.txt 18601203.txt 18811206.txt 19021202.txt 19231206.txt 19450106.txt 19640108.txt 19850206.txt 20050202.txt
17981208.txt 18191207.txt 18401205.txt 18611203.txt 18821204.txt 19031207.txt 19241203.txt 19460121.txt 19650104.txt 19860204.txt 20060131.txt
17991203.txt 18201114.txt 18411207.txt 18621201.txt 18831204.txt 19041206.txt 19251208.txt 19470106.txt 19660112.txt 19870127.txt 20070123.txt
18001111.txt 18211203.txt 18421206.txt 18631208.txt 18841201.txt 19051205.txt 19261207.txt 19480107.txt 19670110.txt 19880125.txt 20080128.txt
18011208.txt 18221203.txt 18431206.txt 18641206.txt 18851208.txt 19061203.txt 19271206.txt 19490105.txt 19680117.txt 19890209.txt 20090224.txt
18021215.txt 18231202.txt 18441203.txt 18651204.txt 18861206.txt 19071203.txt 19281204.txt 19500104.txt 19690114.txt 19900131.txt 20100127.txt
18031017.txt 18241207.txt 18451202.txt 18661203.txt 18871206.txt 19081208.txt 19291203.txt 19510108.txt 19700122.txt 19910129.txt 20110125.txt
18041108.txt 18251206.txt 18461208.txt 18671203.txt 18881203.txt 19091207.txt 19301202.txt 19520109.txt 19710122.txt 19920128.txt 20120124.txt
18051203.txt 18261205.txt 18471207.txt 18681209.txt 18891203.txt 19101206.txt 19311208.txt 19530107.txt 19720120.txt 19930217.txt 20130212.txt
18061202.txt 18271204.txt 18481205.txt 18691206.txt 18901201.txt 19111205.txt 19321206.txt 19530202.txt 19730202.txt 19940125.txt 20140128.txt
18071027.txt 18281202.txt 18491204.txt 18701205.txt 18911209.txt 19121203.txt 19340103.txt 19540107.txt 19740130.txt 19950124.txt 20150120.txt
18081108.txt 18291208.txt 18501202.txt 18711204.txt 18921206.txt 19131202.txt 19350104.txt 19550106.txt 19750115.txt 19960123.txt 20160112.txt
18091129.txt 18301206.txt 18511202.txt 18721202.txt 18931203.txt 19141208.txt 19360103.txt 19560105.txt 19760119.txt 19970204.txt 20170228.txt
raazesh@raazesh-Inspiron-15-7579:~/sou$ vim 20170228.txt
raazesh@raazesh-Inspiron-15-7579:~/sou$ cd ..
raazesh@raazesh-Inspiron-15-7579:~$ tar zcvf sou.tgz sou
$ ls sou.tgz
sou.tgz
```
Raazesh Sainudiin
Thu Aug 31 08:33:38 CEST 2017
# wget http://lamastex.org/datasets/public/SOU/sou.tar.gz
wget http://lamastex.org/datasets/public/SOU/sou.tgz
--2020-09-17 14:09:47-- http://lamastex.org/datasets/public/SOU/sou.tgz
Resolving lamastex.org (lamastex.org)... 166.62.28.100
Connecting to lamastex.org (lamastex.org)|166.62.28.100|:80... connected.
HTTP request sent, awaiting response... 200 OK
Length: 3666765 (3.5M) [application/x-tar]
Saving to: ‘sou.tgz’
0K .......... .......... .......... .......... .......... 1% 132K 27s
50K .......... .......... .......... .......... .......... 2% 266K 20s
100K .......... .......... .......... .......... .......... 4% 269K 17s
150K .......... .......... .......... .......... .......... 5% 2.10M 13s
200K .......... .......... .......... .......... .......... 6% 303K 13s
250K .......... .......... .......... .......... .......... 8% 16.9M 10s
300K .......... .......... .......... .......... .......... 9% 2.34M 9s
350K .......... .......... .......... .......... .......... 11% 302K 9s
400K .......... .......... .......... .......... .......... 12% 2.22M 8s
450K .......... .......... .......... .......... .......... 13% 44.6M 7s
500K .......... .......... .......... .......... .......... 15% 299K 7s
550K .......... .......... .......... .......... .......... 16% 197M 7s
600K .......... .......... .......... .......... .......... 18% 2.48M 6s
650K .......... .......... .......... .......... .......... 19% 50.2M 6s
700K .......... .......... .......... .......... .......... 20% 1.35M 5s
750K .......... .......... .......... .......... .......... 22% 379K 5s
800K .......... .......... .......... .......... .......... 23% 47.7M 5s
850K .......... .......... .......... .......... .......... 25% 2.62M 5s
900K .......... .......... .......... .......... .......... 26% 34.0M 4s
950K .......... .......... .......... .......... .......... 27% 76.1M 4s
1000K .......... .......... .......... .......... .......... 29% 301K 4s
1050K .......... .......... .......... .......... .......... 30% 13.0M 4s
1100K .......... .......... .......... .......... .......... 32% 240M 4s
1150K .......... .......... .......... .......... .......... 33% 3.05M 3s
1200K .......... .......... .......... .......... .......... 34% 29.8M 3s
1250K .......... .......... .......... .......... .......... 36% 44.9M 3s
1300K .......... .......... .......... .......... .......... 37% 49.1M 3s
1350K .......... .......... .......... .......... .......... 39% 302K 3s
1400K .......... .......... .......... .......... .......... 40% 15.4M 3s
1450K .......... .......... .......... .......... .......... 41% 46.0M 3s
1500K .......... .......... .......... .......... .......... 43% 3.36M 2s
1550K .......... .......... .......... .......... .......... 44% 35.9M 2s
1600K .......... .......... .......... .......... .......... 46% 34.6M 2s
1650K .......... .......... .......... .......... .......... 47% 40.9M 2s
1700K .......... .......... .......... .......... .......... 48% 22.9M 2s
1750K .......... .......... .......... .......... .......... 50% 1.55M 2s
1800K .......... .......... .......... .......... .......... 51% 378K 2s
1850K .......... .......... .......... .......... .......... 53% 17.9M 2s
1900K .......... .......... .......... .......... .......... 54% 22.8M 2s
1950K .......... .......... .......... .......... .......... 55% 40.8M 2s
2000K .......... .......... .......... .......... .......... 57% 3.86M 2s
2050K .......... .......... .......... .......... .......... 58% 43.2M 1s
2100K .......... .......... .......... .......... .......... 60% 49.5M 1s
2150K .......... .......... .......... .......... .......... 61% 32.4M 1s
2200K .......... .......... .......... .......... .......... 62% 31.4M 1s
2250K .......... .......... .......... .......... .......... 64% 36.0M 1s
2300K .......... .......... .......... .......... .......... 65% 1.61M 1s
2350K .......... .......... .......... .......... .......... 67% 15.5M 1s
2400K .......... .......... .......... .......... .......... 68% 386K 1s
2450K .......... .......... .......... .......... .......... 69% 15.2M 1s
2500K .......... .......... .......... .......... .......... 71% 46.6M 1s
2550K .......... .......... .......... .......... .......... 72% 47.0M 1s
2600K .......... .......... .......... .......... .......... 74% 6.25M 1s
2650K .......... .......... .......... .......... .......... 75% 7.25M 1s
2700K .......... .......... .......... .......... .......... 76% 53.9M 1s
2750K .......... .......... .......... .......... .......... 78% 15.9M 1s
2800K .......... .......... .......... .......... .......... 79% 22.1M 1s
2850K .......... .......... .......... .......... .......... 80% 144M 1s
2900K .......... .......... .......... .......... .......... 82% 172M 0s
2950K .......... .......... .......... .......... .......... 83% 115M 0s
3000K .......... .......... .......... .......... .......... 85% 53.4M 0s
3050K .......... .......... .......... .......... .......... 86% 1.72M 0s
3100K .......... .......... .......... .......... .......... 87% 21.2M 0s
3150K .......... .......... .......... .......... .......... 89% 377K 0s
3200K .......... .......... .......... .......... .......... 90% 42.5M 0s
3250K .......... .......... .......... .......... .......... 92% 6.89M 0s
3300K .......... .......... .......... .......... .......... 93% 46.7M 0s
3350K .......... .......... .......... .......... .......... 94% 8.75M 0s
3400K .......... .......... .......... .......... .......... 96% 15.4M 0s
3450K .......... .......... .......... .......... .......... 97% 197M 0s
3500K .......... .......... .......... .......... .......... 99% 27.2M 0s
3550K .......... .......... .......... 100% 51.8M=2.5s
2020-09-17 14:09:50 (1.41 MB/s) - ‘sou.tgz’ saved [3666765/3666765]
ls
conf
derby.log
eventlogs
ganglia
logs
sou.tgz
env
MASTER=spark://10.149.224.176:7077
_=/usr/bin/env
LANG=en_US.UTF-8
SUDO_GID=0
OLDPWD=/databricks/chauffeur
PTY_LIB_FOLDER=/usr/lib/libpty
HIVE_HOME=/home/ubuntu/hive-0.9.0-bin
JAVA_OPTS= -Djava.io.tmpdir=/local_disk0/tmp -XX:MaxPermSize=512m -XX:-OmitStackTraceInFastThrow -Djava.security.properties=/databricks/spark/dbconf/java/extra.security -XX:+PrintFlagsFinal -XX:+PrintGCDateStamps -verbose:gc -XX:+PrintGCDetails -Xss4m -Djavax.xml.datatype.DatatypeFactory=com.sun.org.apache.xerces.internal.jaxp.datatype.DatatypeFactoryImpl -Djavax.xml.parsers.DocumentBuilderFactory=com.sun.org.apache.xerces.internal.jaxp.DocumentBuilderFactoryImpl -Djavax.xml.parsers.SAXParserFactory=com.sun.org.apache.xerces.internal.jaxp.SAXParserFactoryImpl -Djavax.xml.validation.SchemaFactory:http://www.w3.org/2001/XMLSchema=com.sun.org.apache.xerces.internal.jaxp.validation.XMLSchemaFactory -Dorg.xml.sax.driver=com.sun.org.apache.xerces.internal.parsers.SAXParser -Dorg.w3c.dom.DOMImplementationSourceList=com.sun.org.apache.xerces.internal.dom.DOMXSImplementationSourceImpl -Djavax.net.ssl.sessionCacheSize=10000 -Dscala.reflect.runtime.disable.typetag.cache=true -Ddatabricks.serviceName=driver-1 -Xms1492m -Xmx1492m -Dspark.ui.port=47839 -Dspark.executor.extraJavaOptions="-Djava.io.tmpdir=/local_disk0/tmp -XX:ReservedCodeCacheSize=256m -XX:+UseCodeCacheFlushing -Djava.security.properties=/databricks/spark/dbconf/java/extra.security -XX:+PrintFlagsFinal -XX:+PrintGCDateStamps -verbose:gc -XX:+PrintGCDetails -Xss4m -Djavax.xml.datatype.DatatypeFactory=com.sun.org.apache.xerces.internal.jaxp.datatype.DatatypeFactoryImpl -Djavax.xml.parsers.DocumentBuilderFactory=com.sun.org.apache.xerces.internal.jaxp.DocumentBuilderFactoryImpl -Djavax.xml.parsers.SAXParserFactory=com.sun.org.apache.xerces.internal.jaxp.SAXParserFactoryImpl -Djavax.xml.validation.SchemaFactory:http://www.w3.org/2001/XMLSchema=com.sun.org.apache.xerces.internal.jaxp.validation.XMLSchemaFactory -Dorg.xml.sax.driver=com.sun.org.apache.xerces.internal.parsers.SAXParser -Dorg.w3c.dom.DOMImplementationSourceList=com.sun.org.apache.xerces.internal.dom.DOMXSImplementationSourceImpl -Djavax.net.ssl.sessionCacheSize=10000 -Dscala.reflect.runtime.disable.typetag.cache=true -Ddatabricks.serviceName=spark-executor-1" -Dspark.executor.memory=2516m -Dspark.executor.extraClassPath=/databricks/spark/dbconf/log4j/executor:/databricks/spark/dbconf/jets3t/:/databricks/spark/dbconf/hadoop:/databricks/hive/conf:/databricks/jars/api-base--api-base_java-spark_3.0_2.12_deploy.jar:/databricks/jars/api-base--api-base-spark_3.0_2.12_deploy.jar:/databricks/jars/api--rpc--rpc_parser-spark_3.0_2.12_deploy.jar:/databricks/jars/chauffeur-api--api--endpoints--endpoints-spark_3.0_2.12_deploy.jar:/databricks/jars/chauffeur-api--chauffeur-api-spark_3.0_2.12_deploy.jar:/databricks/jars/common--client--client-spark_3.0_2.12_deploy.jar:/databricks/jars/common--common-spark_3.0_2.12_deploy.jar:/databricks/jars/common--credentials--credentials-spark_3.0_2.12_deploy.jar:/databricks/jars/common--hadoop--hadoop-spark_3.0_2.12_deploy.jar:/databricks/jars/common--java-flight-recorder--java-flight-recorder-spark_3.0_2.12_deploy.jar:/databricks/jars/common--jetty--client--client-spark_3.0_2.12_deploy.jar:/databricks/jars/common--lazy--lazy-spark_3.0_2.12_deploy.jar:/databricks/jars/common--libcommon_resources.jar:/databricks/jars/common--path--path-spark_3.0_2.12_deploy.jar:/databricks/jars/common--rate-limiter--rate-limiter-spark_3.0_2.12_deploy.jar:/databricks/jars/common--storage--storage-spark_3.0_2.12_deploy.jar:/databricks/jars/common--tracing--tracing-spark_3.0_2.12_deploy.jar:/databricks/jars/common--util--locks-spark_3.0_2.12_deploy.jar:/databricks/jars/daemon--data--client--client-spark_3.0_2.12_deploy.jar:/databricks/jars/daemon--data--client--conf--conf-spark_3.0_2.12_deploy.jar:/databricks/jars/daemon--data--client--utils-spark_3.0_2.12_deploy.jar:/databricks/jars/daemon--data--data-common--data-common-spark_3.0_2.12_deploy.jar:/databricks/jars/dbfs--exceptions--exceptions-spark_3.0_2.12_deploy.jar:/databricks/jars/dbfs--utils--dbfs-utils-spark_3.0_2.12_deploy.jar:/databricks/jars/extern--acl--auth--auth-spark_3.0_2.12_deploy.jar:/databricks/jars/extern--extern-spark_3.0_2.12_deploy.jar:/databricks/jars/extern--libaws-regions.jar:/databricks/jars/----jackson_annotations_shaded--libjackson-annotations.jar:/databricks/jars/----jackson_core_shaded--libjackson-core.jar:/databricks/jars/----jackson_databind_shaded--libjackson-databind.jar:/databricks/jars/----jackson_datatype_joda_shaded--libjackson-datatype-joda.jar:/databricks/jars/jsonutil--jsonutil-spark_3.0_2.12_deploy.jar:/databricks/jars/libraries--api--managedLibraries--managedLibraries-spark_3.0_2.12_deploy.jar:/databricks/jars/libraries--api--typemappers--typemappers-spark_3.0_2.12_deploy.jar:/databricks/jars/libraries--libraries-spark_3.0_2.12_deploy.jar:/databricks/jars/logging--log4j-mod--log4j-mod-spark_3.0_2.12_deploy.jar:/databricks/jars/logging--utils--logging-utils-spark_3.0_2.12_deploy.jar:/databricks/jars/s3commit--client--client-spark_3.0_2.12_deploy.jar:/databricks/jars/s3commit--common--common-spark_3.0_2.12_deploy.jar:/databricks/jars/s3--s3-spark_3.0_2.12_deploy.jar:/databricks/jars/----scalapb_090--com.lihaoyi__fastparse_2.12__2.1.3_shaded.jar:/databricks/jars/----scalapb_090--com.lihaoyi__sourcecode_2.12__0.1.7_shaded.jar:/databricks/jars/----scalapb_090--runtime-unshaded-jetty9-hadoop1_2.12_deploy_shaded.jar:/databricks/jars/secret-manager--api--api-spark_3.0_2.12_deploy.jar:/databricks/jars/spark--common--spark-common-spark_3.0_2.12_deploy.jar:/databricks/jars/spark--common-utils--utils-spark_3.0_2.12_deploy.jar:/databricks/jars/spark--conf-reader--conf-reader_lib-spark_3.0_2.12_deploy.jar:/databricks/jars/spark--dbutils--dbutils-api-spark_3.0_2.12_deploy.jar:/databricks/jars/spark--driver--antlr--parser-spark_3.0_2.12_deploy.jar:/databricks/jars/spark--driver--common--driver-common-spark_3.0_2.12_deploy.jar:/databricks/jars/spark--driver--display--display-spark_3.0_2.12_deploy.jar:/databricks/jars/spark--driver--driver-spark_3.0_2.12_deploy.jar:/databricks/jars/spark--driver--events-spark_3.0_2.12_deploy.jar:/databricks/jars/spark--driver--ml--ml-spark_3.0_2.12_deploy.jar:/databricks/jars/spark--driver--secret-redaction-spark_3.0_2.12_deploy.jar:/databricks/jars/spark--driver--spark--resources-resources.jar:/databricks/jars/spark--sql-extension--sql-extension-spark_3.0_2.12_deploy.jar:/databricks/jars/spark--versions--3.0--shim_2.12_deploy.jar:/databricks/jars/spark--versions--3.0--spark_2.12_deploy.jar:/databricks/jars/sqlgateway--common--endpoint_id-spark_3.0_2.12_deploy.jar:/databricks/jars/sqlgateway--history--api--api-spark_3.0_2.12_deploy.jar:/databricks/jars/third_party--armeria--maven-trees_armeria_com.fasterxml.jackson.core_jackson-annotations_com.fasterxml.jackson.core__jackson-annotations__2.11.1_shaded.jar:/databricks/jars/third_party--armeria--maven-trees_armeria_com.fasterxml.jackson.core_jackson-core_com.fasterxml.jackson.core__jackson-core__2.11.1_shaded.jar:/databricks/jars/third_party--armeria--maven-trees_armeria_com.fasterxml.jackson.core_jackson-databind_com.fasterxml.jackson.core__jackson-databind__2.11.1_shaded.jar:/databricks/jars/third_party--armeria--maven-trees_armeria_com.google.android_annotations_com.google.android__annotations__4.1.1.4_shaded.jar:/databricks/jars/third_party--armeria--maven-trees_armeria_com.google.api.grpc_proto-google-common-protos_com.google.api.grpc__proto-google-common-protos__1.17.0_shaded.jar:/databricks/jars/third_party--armeria--maven-trees_armeria_com.google.code.findbugs_jsr305_com.google.code.findbugs__jsr305__3.0.2_shaded.jar:/databricks/jars/third_party--armeria--maven-trees_armeria_com.google.code.gson_gson_com.google.code.gson__gson__2.8.6_shaded.jar:/databricks/jars/third_party--armeria--maven-trees_armeria_com.google.errorprone_error_prone_annotations_com.google.errorprone__error_prone_annotations__2.3.4_shaded.jar:/databricks/jars/third_party--armeria--maven-trees_armeria_com.google.guava_failureaccess_com.google.guava__failureaccess__1.0.1_shaded.jar:/databricks/jars/third_party--armeria--maven-trees_armeria_com.google.guava_guava_com.google.guava__guava__28.2-android_shaded.jar:/databricks/jars/third_party--armeria--maven-trees_armeria_com.google.guava_listenablefuture_com.google.guava__listenablefuture__9999.0-empty-to-avoid-conflict-with-guava_shaded.jar:/databricks/jars/third_party--armeria--maven-trees_armeria_com.google.j2objc_j2objc-annotations_com.google.j2objc__j2objc-annotations__1.3_shaded.jar:/databricks/jars/third_party--armeria--maven-trees_armeria_com.google.protobuf_protobuf-java_com.google.protobuf__protobuf-java__3.12.0_shaded.jar:/databricks/jars/third_party--armeria--maven-trees_armeria_com.google.protobuf_protobuf-java-util_com.google.protobuf__protobuf-java-util__3.12.0_shaded.jar:/databricks/jars/third_party--armeria--maven-trees_armeria_com.linecorp.armeria_armeria-brave_com.linecorp.armeria__armeria-brave__0.99.8_shaded.jar:/databricks/jars/third_party--armeria--maven-trees_armeria_com.linecorp.armeria_armeria_com.linecorp.armeria__armeria__0.99.8_shaded.jar:/databricks/jars/third_party--armeria--maven-trees_armeria_com.linecorp.armeria_armeria-grpc_com.linecorp.armeria__armeria-grpc__0.99.8_shaded.jar:/databricks/jars/third_party--armeria--maven-trees_armeria_com.linecorp.armeria_armeria-grpc-protocol_com.linecorp.armeria__armeria-grpc-protocol__0.99.8_shaded.jar:/databricks/jars/third_party--armeria--maven-trees_armeria_io.grpc_grpc-api_io.grpc__grpc-api__1.30.2_shaded.jar:/databricks/jars/third_party--armeria--maven-trees_armeria_io.grpc_grpc-context_io.grpc__grpc-context__1.30.2_shaded.jar:/databricks/jars/third_party--armeria--maven-trees_armeria_io.grpc_grpc-core_io.grpc__grpc-core__1.30.2_shaded.jar:/databricks/jars/third_party--armeria--maven-trees_armeria_io.grpc_grpc-protobuf_io.grpc__grpc-protobuf__1.30.2_shaded.jar:/databricks/jars/third_party--armeria--maven-trees_armeria_io.grpc_grpc-protobuf-lite_io.grpc__grpc-protobuf-lite__1.30.2_shaded.jar:/databricks/jars/third_party--armeria--maven-trees_armeria_io.grpc_grpc-services_io.grpc__grpc-services__1.30.2_shaded.jar:/databricks/jars/third_party--armeria--maven-trees_armeria_io.grpc_grpc-stub_io.grpc__grpc-stub__1.30.2_shaded.jar:/databricks/jars/third_party--armeria--maven-trees_armeria_io.micrometer_micrometer-core_io.micrometer__micrometer-core__1.5.2_shaded.jar:/databricks/jars/third_party--armeria--maven-trees_armeria_io.netty_netty-buffer_io.netty__netty-buffer__4.1.50.Final_shaded.jar:/databricks/jars/third_party--armeria--maven-trees_armeria_io.netty_netty-codec-dns_io.netty__netty-codec-dns__4.1.50.Final_shaded.jar:/databricks/jars/third_party--armeria--maven-trees_armeria_io.netty_netty-codec-haproxy_io.netty__netty-codec-haproxy__4.1.50.Final_shaded.jar:/databricks/jars/third_party--armeria--maven-trees_armeria_io.netty_netty-codec-http2_io.netty__netty-codec-http2__4.1.50.Final_shaded.jar:/databricks/jars/third_party--armeria--maven-trees_armeria_io.netty_netty-codec-http_io.netty__netty-codec-http__4.1.50.Final_shaded.jar:/databricks/jars/third_party--armeria--maven-trees_armeria_io.netty_netty-codec_io.netty__netty-codec__4.1.50.Final_shaded.jar:/databricks/jars/third_party--armeria--maven-trees_armeria_io.netty_netty-codec-socks_io.netty__netty-codec-socks__4.1.50.Final_shaded.jar:/databricks/jars/third_party--armeria--maven-trees_armeria_io.netty_netty-common_io.netty__netty-common__4.1.50.Final_shaded.jar:/databricks/jars/third_party--armeria--maven-trees_armeria_io.netty_netty-handler_io.netty__netty-handler__4.1.50.Final_shaded.jar:/databricks/jars/third_party--armeria--maven-trees_armeria_io.netty_netty-handler-proxy_io.netty__netty-handler-proxy__4.1.50.Final_shaded.jar:/databricks/jars/third_party--armeria--maven-trees_armeria_io.netty_netty-resolver-dns_io.netty__netty-resolver-dns__4.1.50.Final_shaded.jar:/databricks/jars/third_party--armeria--maven-trees_armeria_io.netty_netty-resolver_io.netty__netty-resolver__4.1.50.Final_shaded.jar:/databricks/jars/third_party--armeria--maven-trees_armeria_io.netty_netty-tcnative-boringssl-static_io.netty__netty-tcnative-boringssl-static__2.0.30.Final_shaded.jar:/databricks/jars/third_party--armeria--maven-trees_armeria_io.netty_netty-transport_io.netty__netty-transport__4.1.50.Final_shaded.jar:/databricks/jars/third_party--armeria--maven-trees_armeria_io.netty_netty-transport-native-epoll-linux-x86_64_io.netty__netty-transport-native-epoll-linux-x86_64__4.1.50.Final_shaded.jar:/databricks/jars/third_party--armeria--maven-trees_armeria_io.netty_netty-transport-native-unix-common_io.netty__netty-transport-native-unix-common__4.1.50.Final_shaded.jar:/databricks/jars/third_party--armeria--maven-trees_armeria_io.netty_netty-transport-native-unix-common-linux-x86_64_io.netty__netty-transport-native-unix-common-linux-x86_64__4.1.50.Final_shaded.jar:/databricks/jars/third_party--armeria--maven-trees_armeria_io.perfmark_perfmark-api_io.perfmark__perfmark-api__0.19.0_shaded.jar:/databricks/jars/third_party--armeria--maven-trees_armeria_io.zipkin.brave_brave-instrumentation-http_io.zipkin.brave__brave-instrumentation-http__5.12.3_shaded.jar:/databricks/jars/third_party--armeria--maven-trees_armeria_io.zipkin.brave_brave_io.zipkin.brave__brave__5.12.3_shaded.jar:/databricks/jars/third_party--armeria--maven-trees_armeria_io.zipkin.reporter2_zipkin-reporter-brave_io.zipkin.reporter2__zipkin-reporter-brave__2.15.0_shaded.jar:/databricks/jars/third_party--armeria--maven-trees_armeria_io.zipkin.reporter2_zipkin-reporter_io.zipkin.reporter2__zipkin-reporter__2.15.0_shaded.jar:/databricks/jars/third_party--armeria--maven-trees_armeria_io.zipkin.zipkin2_zipkin_io.zipkin.zipkin2__zipkin__2.21.1_shaded.jar:/databricks/jars/third_party--armeria--maven-trees_armeria_jakarta.annotation_jakarta.annotation-api_jakarta.annotation__jakarta.annotation-api__1.3.5_shaded.jar:/databricks/jars/third_party--armeria--maven-trees_armeria_liball_deps_2.12_shaded.jar:/databricks/jars/third_party--armeria--maven-trees_armeria_net.bytebuddy_byte-buddy_net.bytebuddy__byte-buddy__1.10.9_shaded.jar:/databricks/jars/third_party--armeria--maven-trees_armeria_org.checkerframework_checker-compat-qual_org.checkerframework__checker-compat-qual__2.5.5_shaded.jar:/databricks/jars/third_party--armeria--maven-trees_armeria_org.codehaus.mojo_animal-sniffer-annotations_org.codehaus.mojo__animal-sniffer-annotations__1.18_shaded.jar:/databricks/jars/third_party--armeria--maven-trees_armeria_org.curioswitch.curiostack_protobuf-jackson_org.curioswitch.curiostack__protobuf-jackson__1.1.0_shaded.jar:/databricks/jars/third_party--armeria--maven-trees_armeria_org.hdrhistogram_HdrHistogram_org.hdrhistogram__HdrHistogram__2.1.12_shaded.jar:/databricks/jars/third_party--armeria--maven-trees_armeria_org.joda_joda-convert_org.joda__joda-convert__2.2.1_shaded.jar:/databricks/jars/third_party--armeria--maven-trees_armeria_org.latencyutils_LatencyUtils_org.latencyutils__LatencyUtils__2.0.3_shaded.jar:/databricks/jars/third_party--armeria--maven-trees_armeria_org.reactivestreams_reactive-streams_org.reactivestreams__reactive-streams__1.0.3_shaded.jar:/databricks/jars/third_party--azure--com.fasterxml.jackson.core__jackson-core__2.7.2_shaded.jar:/databricks/jars/third_party--azure--com.microsoft.azure__azure-client-runtime__1.7.0_container_shaded.jar:/databricks/jars/third_party--azure--com.microsoft.azure__azure-keyvault-core__1.0.0_shaded.jar:/databricks/jars/third_party--azure--com.microsoft.azure__azure-storage__8.6.4_shaded.jar:/databricks/jars/third_party--azure--com.microsoft.rest__client-runtime__1.7.0_container_shaded.jar:/databricks/jars/third_party--azure--org.apache.commons__commons-lang3__3.4_shaded.jar:/databricks/jars/third_party--datalake--datalake-spark_3.0_2.12_deploy.jar:/databricks/jars/third_party--dropwizard-metrics-log4j-v3.2.6--metrics-log4j-spark_3.0_2.12_deploy.jar:/databricks/jars/third_party--gcs--animal-sniffer-annotations_shaded.jar:/databricks/jars/third_party--gcs--annotations_shaded.jar:/databricks/jars/third_party--gcs--api-common_shaded.jar:/databricks/jars/third_party--gcs--auto-value-annotations_shaded.jar:/databricks/jars/third_party--gcs--checker-compat-qual_shaded.jar:/databricks/jars/third_party--gcs--checker-qual_shaded.jar:/databricks/jars/third_party--gcs--commons-codec_shaded.jar:/databricks/jars/third_party--gcs--commons-lang3_shaded.jar:/databricks/jars/third_party--gcs--commons-logging_shaded.jar:/databricks/jars/third_party--gcs--conscrypt-openjdk-uber_shaded.jar:/databricks/jars/third_party--gcs--error_prone_annotations_shaded.jar:/databricks/jars/third_party--gcs--failureaccess_shaded.jar:/databricks/jars/third_party--gcs--flogger_shaded.jar:/databricks/jars/third_party--gcs--flogger-slf4j-backend_shaded.jar:/databricks/jars/third_party--gcs--flogger-system-backend_shaded.jar:/databricks/jars/third_party--gcs--gcs-connector_shaded.jar:/databricks/jars/third_party--gcs--gcsio_shaded.jar:/databricks/jars/third_party--gcs--gcs-shaded-jetty9-hadoop1_2.12_deploy.jar:/databricks/jars/third_party--gcs--google-api-client-jackson2_shaded.jar:/databricks/jars/third_party--gcs--google-api-client-java6_shaded.jar:/databricks/jars/third_party--gcs--google-api-client_shaded.jar:/databricks/jars/third_party--gcs--google-api-services-storage_shaded.jar:/databricks/jars/third_party--gcs--google-auth-library-credentials_shaded.jar:/databricks/jars/third_party--gcs--google-auth-library-oauth2-http_shaded.jar:/databricks/jars/third_party--gcs--google-extensions_shaded.jar:/databricks/jars/third_party--gcs--google-http-client-jackson2_shaded.jar:/databricks/jars/third_party--gcs--google-http-client_shaded.jar:/databricks/jars/third_party--gcs--google-oauth-client-java6_shaded.jar:/databricks/jars/third_party--gcs--google-oauth-client_shaded.jar:/databricks/jars/third_party--gcs--grpc-alts_shaded.jar:/databricks/jars/third_party--gcs--grpc-api_shaded.jar:/databricks/jars/third_party--gcs--grpc-auth_shaded.jar:/databricks/jars/third_party--gcs--grpc-context_shaded.jar:/databricks/jars/third_party--gcs--grpc-core_shaded.jar:/databricks/jars/third_party--gcs--grpc-grpclb_shaded.jar:/databricks/jars/third_party--gcs--grpc-netty-shaded_shaded.jar:/databricks/jars/third_party--gcs--grpc-protobuf-lite_shaded.jar:/databricks/jars/third_party--gcs--grpc-protobuf_shaded.jar:/databricks/jars/third_party--gcs--grpc-stub_shaded.jar:/databricks/jars/third_party--gcs--gson_shaded.jar:/databricks/jars/third_party--gcs--guava_shaded.jar:/databricks/jars/third_party--gcs--httpclient_shaded.jar:/databricks/jars/third_party--gcs--httpcore_shaded.jar:/databricks/jars/third_party--gcs--j2objc-annotations_shaded.jar:/databricks/jars/third_party--gcs--jackson-core_shaded.jar:/databricks/jars/third_party--gcs--javax.annotation-api_shaded.jar:/databricks/jars/third_party--gcs--jsr305_shaded.jar:/databricks/jars/third_party--gcs--listenablefuture_shaded.jar:/databricks/jars/third_party--gcs--opencensus-api_shaded.jar:/databricks/jars/third_party--gcs--opencensus-contrib-http-util_shaded.jar:/databricks/jars/third_party--gcs--perfmark-api_shaded.jar:/databricks/jars/third_party--gcs--protobuf-java_shaded.jar:/databricks/jars/third_party--gcs--protobuf-java-util_shaded.jar:/databricks/jars/third_party--gcs--proto-google-common-protos_shaded.jar:/databricks/jars/third_party--gcs--proto-google-iam-v1_shaded.jar:/databricks/jars/third_party--gcs--util-hadoop_shaded.jar:/databricks/jars/third_party--gcs--util_shaded.jar:/databricks/jars/third_party--hadoop-azure-2.7.3-abfs--aopalliance__aopalliance__1.0_2.12_shaded_20180625_3682417_spark_3.0.jar:/databricks/jars/third_party--hadoop-azure-2.7.3-abfs--com.fasterxml.jackson.core__jackson-annotations__2.7.0_2.12_shaded_20180625_3682417_spark_3.0.jar:/databricks/jars/third_party--hadoop-azure-2.7.3-abfs--com.fasterxml.jackson.core__jackson-core__2.7.2_2.12_shaded_20180625_3682417_spark_3.0.jar:/databricks/jars/third_party--hadoop-azure-2.7.3-abfs--com.fasterxml.jackson.core__jackson-core__2.7.2_2.12_shaded_20180920_b33d810_spark_3.0.jar:/databricks/jars/third_party--hadoop-azure-2.7.3-abfs--com.fasterxml.jackson.core__jackson-databind__2.7.2_2.12_shaded_20180625_3682417_spark_3.0.jar:/databricks/jars/third_party--hadoop-azure-2.7.3-abfs--com.fasterxml.jackson.datatype__jackson-datatype-joda__2.7.2_2.12_shaded_20180625_3682417_spark_3.0.jar:/databricks/jars/third_party--hadoop-azure-2.7.3-abfs--com.google.code.findbugs__jsr305__1.3.9_2.12_shaded_20180920_b33d810_spark_3.0.jar:/databricks/jars/third_party--hadoop-azure-2.7.3-abfs--com.google.guava__guava__11.0.2_2.12_shaded_20180920_b33d810_spark_3.0.jar:/databricks/jars/third_party--hadoop-azure-2.7.3-abfs--com.google.guava__guava__16.0_2.12_shaded_20180625_3682417_spark_3.0.jar:/databricks/jars/third_party--hadoop-azure-2.7.3-abfs--com.google.inject__guice__3.0_2.12_shaded_20180625_3682417_spark_3.0.jar:/databricks/jars/third_party--hadoop-azure-2.7.3-abfs--com.microsoft.azure__azure-annotations__1.2.0_2.12_shaded_20180625_3682417_spark_3.0.jar:/databricks/jars/third_party--hadoop-azure-2.7.3-abfs--com.microsoft.azure__azure-keyvault-core__1.0.0_2.12_shaded_20180625_3682417_spark_3.0.jar:/databricks/jars/third_party--hadoop-azure-2.7.3-abfs--com.microsoft.azure__azure-keyvault-core__1.0.0_2.12_shaded_20180920_b33d810_spark_3.0.jar:/databricks/jars/third_party--hadoop-azure-2.7.3-abfs--com.microsoft.azure__azure-storage__7.0.0_2.12_shaded_20180920_b33d810_spark_3.0.jar:/databricks/jars/third_party--hadoop-azure-2.7.3-abfs--com.microsoft.azure__azure-storage__8.6.4_2.12_shaded_20180625_3682417_spark_3.0.jar:/databricks/jars/third_party--hadoop-azure-2.7.3-abfs--com.microsoft.rest__client-runtime__1.1.0_2.12_shaded_20180625_3682417_spark_3.0.jar:/databricks/jars/third_party--hadoop-azure-2.7.3-abfs--commons-codec__commons-codec__1.9_2.12_shaded_20180625_3682417_spark_3.0.jar:/databricks/jars/third_party--hadoop-azure-2.7.3-abfs--commons-codec__commons-codec__1.9_2.12_shaded_20180920_b33d810_spark_3.0.jar:/databricks/jars/third_party--hadoop-azure-2.7.3-abfs--commons-logging__commons-logging__1.2_2.12_shaded_20180625_3682417_spark_3.0.jar:/databricks/jars/third_party--hadoop-azure-2.7.3-abfs--commons-logging__commons-logging__1.2_2.12_shaded_20180920_b33d810_spark_3.0.jar:/databricks/jars/third_party--hadoop-azure-2.7.3-abfs--com.squareup.okhttp3__logging-interceptor__3.3.1_2.12_shaded_20180625_3682417_spark_3.0.jar:/databricks/jars/third_party--hadoop-azure-2.7.3-abfs--com.squareup.okhttp3__okhttp__3.3.1_2.12_shaded_20180625_3682417_spark_3.0.jar:/databricks/jars/third_party--hadoop-azure-2.7.3-abfs--com.squareup.okhttp3__okhttp-urlconnection__3.3.1_2.12_shaded_20180625_3682417_spark_3.0.jar:/databricks/jars/third_party--hadoop-azure-2.7.3-abfs--com.squareup.okio__okio__1.8.0_2.12_shaded_20180625_3682417_spark_3.0.jar:/databricks/jars/third_party--hadoop-azure-2.7.3-abfs--com.squareup.retrofit2__adapter-rxjava__2.1.0_2.12_shaded_20180625_3682417_spark_3.0.jar:/databricks/jars/third_party--hadoop-azure-2.7.3-abfs--com.squareup.retrofit2__converter-jackson__2.1.0_2.12_shaded_20180625_3682417_spark_3.0.jar:/databricks/jars/third_party--hadoop-azure-2.7.3-abfs--com.squareup.retrofit2__retrofit__2.1.0_2.12_shaded_20180625_3682417_spark_3.0.jar:/databricks/jars/third_party--hadoop-azure-2.7.3-abfs--com.sun.xml.bind__jaxb-impl__2.2.3-1_2.12_shaded_20180625_3682417_spark_3.0.jar:/databricks/jars/third_party--hadoop-azure-2.7.3-abfs--hadoop-azure-2.7.3-abfs-external-20180625_3682417-spark_3.0_2.12_deploy_shaded.jar:/databricks/jars/third_party--hadoop-azure-2.7.3-abfs--hadoop-azure-2.7.3-abfs-external-20180920_b33d810-spark_3.0_2.12_deploy_shaded.jar:/databricks/jars/third_party--hadoop-azure-2.7.3-abfs--io.netty__netty-all__4.0.52.Final_2.12_shaded_20180625_3682417_spark_3.0.jar:/databricks/jars/third_party--hadoop-azure-2.7.3-abfs--io.reactivex__rxjava__1.2.4_2.12_shaded_20180625_3682417_spark_3.0.jar:/databricks/jars/third_party--hadoop-azure-2.7.3-abfs--javax.activation__activation__1.1_2.12_shaded_20180625_3682417_spark_3.0.jar:/databricks/jars/third_party--hadoop-azure-2.7.3-abfs--javax.inject__javax.inject__1_2.12_shaded_20180625_3682417_spark_3.0.jar:/databricks/jars/third_party--hadoop-azure-2.7.3-abfs--javax.xml.bind__jaxb-api__2.2.2_2.12_shaded_20180625_3682417_spark_3.0.jar:/databricks/jars/third_party--hadoop-azure-2.7.3-abfs--javax.xml.stream__stax-api__1.0-2_2.12_shaded_20180625_3682417_spark_3.0.jar:/databricks/jars/third_party--hadoop-azure-2.7.3-abfs--joda-time__joda-time__2.4_2.12_shaded_20180625_3682417_spark_3.0.jar:/databricks/jars/third_party--hadoop-azure-2.7.3-abfs--org.apache.htrace__htrace-core__3.1.0-incubating_2.12_shaded_20180625_3682417_spark_3.0.jar:/databricks/jars/third_party--hadoop-azure-2.7.3-abfs--org.apache.httpcomponents__httpclient__4.5.2_2.12_shaded_20180625_3682417_spark_3.0.jar:/databricks/jars/third_party--hadoop-azure-2.7.3-abfs--org.apache.httpcomponents__httpclient__4.5.2_2.12_shaded_20180920_b33d810_spark_3.0.jar:/databricks/jars/third_party--hadoop-azure-2.7.3-abfs--org.apache.httpcomponents__httpcore__4.4.4_2.12_shaded_20180625_3682417_spark_3.0.jar:/databricks/jars/third_party--hadoop-azure-2.7.3-abfs--org.apache.httpcomponents__httpcore__4.4.4_2.12_shaded_20180920_b33d810_spark_3.0.jar:/databricks/jars/third_party--hadoop--hadoop-tools--hadoop-aws--lib-spark_3.0_2.12_deploy_shaded.jar:/databricks/jars/third_party--jackson--guava_only_shaded.jar:/databricks/jars/third_party--jackson--jackson-module-scala-shaded_2.12_deploy.jar:/databricks/jars/third_party--jackson--jsr305_only_shaded.jar:/databricks/jars/third_party--jackson--paranamer_only_shaded.jar:/databricks/jars/third_party--jetty8-shaded-client--databricks-patched-jetty-client-jar_shaded.jar:/databricks/jars/third_party--jetty8-shaded-client--databricks-patched-jetty-http-jar_shaded.jar:/databricks/jars/third_party--jetty8-shaded-client--jetty-io_shaded.jar:/databricks/jars/third_party--jetty8-shaded-client--jetty-jmx_shaded.jar:/databricks/jars/third_party--jetty8-shaded-client--jetty-util_shaded.jar:/databricks/jars/third_party--jetty-client--jetty-client_shaded.jar:/databricks/jars/third_party--jetty-client--jetty-http_shaded.jar:/databricks/jars/third_party--jetty-client--jetty-io_shaded.jar:/databricks/jars/third_party--jetty-client--jetty-util_shaded.jar:/databricks/jars/third_party--opencensus-shaded--com.google.code.findbugs__jsr305__3.0.2_shaded.jar:/databricks/jars/third_party--opencensus-shaded--com.google.code.gson__gson__2.8.2_shaded.jar:/databricks/jars/third_party--opencensus-shaded--com.google.errorprone__error_prone_annotations__2.1.3_shaded.jar:/databricks/jars/third_party--opencensus-shaded--com.google.guava__guava__26.0-android_shaded.jar:/databricks/jars/third_party--opencensus-shaded--com.google.j2objc__j2objc-annotations__1.1_shaded.jar:/databricks/jars/third_party--opencensus-shaded--com.lmax__disruptor__3.4.2_shaded.jar:/databricks/jars/third_party--opencensus-shaded--commons-codec__commons-codec__1.9_shaded.jar:/databricks/jars/third_party--opencensus-shaded--commons-logging__commons-logging__1.2_shaded.jar:/databricks/jars/third_party--opencensus-shaded--com.squareup.okhttp3__okhttp__3.9.0_shaded.jar:/databricks/jars/third_party--opencensus-shaded--com.squareup.okio__okio__1.13.0_shaded.jar:/databricks/jars/third_party--opencensus-shaded--io.grpc__grpc-context__1.19.0_shaded.jar:/databricks/jars/third_party--opencensus-shaded--io.jaegertracing__jaeger-client__0.33.1_shaded.jar:/databricks/jars/third_party--opencensus-shaded--io.jaegertracing__jaeger-core__0.33.1_shaded.jar:/databricks/jars/third_party--opencensus-shaded--io.jaegertracing__jaeger-thrift__0.33.1_shaded.jar:/databricks/jars/third_party--opencensus-shaded--io.jaegertracing__jaeger-tracerresolver__0.33.1_shaded.jar:/databricks/jars/third_party--opencensus-shaded--io.opencensus__opencensus-api__0.22.1_shaded.jar:/databricks/jars/third_party--opencensus-shaded--io.opencensus__opencensus-exporter-trace-jaeger__0.22.1_shaded.jar:/databricks/jars/third_party--opencensus-shaded--io.opencensus__opencensus-exporter-trace-util__0.22.1_shaded.jar:/databricks/jars/third_party--opencensus-shaded--io.opencensus__opencensus-impl__0.22.1_shaded.jar:/databricks/jars/third_party--opencensus-shaded--io.opencensus__opencensus-impl-core__0.22.1_shaded.jar:/databricks/jars/third_party--opencensus-shaded--io.opentracing.contrib__opentracing-tracerresolver__0.1.5_shaded.jar:/databricks/jars/third_party--opencensus-shaded--io.opentracing__opentracing-api__0.31.0_shaded.jar:/databricks/jars/third_party--opencensus-shaded--io.opentracing__opentracing-noop__0.31.0_shaded.jar:/databricks/jars/third_party--opencensus-shaded--io.opentracing__opentracing-util__0.31.0_shaded.jar:/databricks/jars/third_party--opencensus-shaded--org.apache.httpcomponents__httpclient__4.4.1_shaded.jar:/databricks/jars/third_party--opencensus-shaded--org.apache.httpcomponents__httpcore__4.4.1_shaded.jar:/databricks/jars/third_party--opencensus-shaded--org.apache.thrift__libthrift__0.11.0_shaded.jar:/databricks/jars/third_party--opencensus-shaded--org.checkerframework__checker-compat-qual__2.5.2_shaded.jar:/databricks/jars/third_party--opencensus-shaded--org.codehaus.mojo__animal-sniffer-annotations__1.14_shaded.jar:/databricks/jars/third_party--prometheus-client--jmx_collector-spark_3.0_2.12_deploy.jar:/databricks/jars/third_party--prometheus-client--simpleclient_common-spark_3.0_2.12_deploy.jar:/databricks/jars/third_party--prometheus-client--simpleclient_dropwizard-spark_3.0_2.12_deploy.jar:/databricks/jars/third_party--prometheus-client--simpleclient_servlet-spark_3.0_2.12_deploy.jar:/databricks/jars/third_party--prometheus-client--simpleclient-spark_3.0_2.12_deploy.jar:/databricks/jars/utils--process_utils-spark_3.0_2.12_deploy.jar:/databricks/jars/workflow--workflow-spark_3.0_2.12_deploy.jar:/databricks/jars/----workspace_spark_3_0--common--kvstore--kvstore-hive-2.3__hadoop-2.7_2.12_deploy.jar:/databricks/jars/----workspace_spark_3_0--common--network-common--network-common-hive-2.3__hadoop-2.7_2.12_deploy.jar:/databricks/jars/----workspace_spark_3_0--common--network-shuffle--network-shuffle-hive-2.3__hadoop-2.7_2.12_deploy.jar:/databricks/jars/----workspace_spark_3_0--common--sketch--sketch-hive-2.3__hadoop-2.7_2.12_deploy.jar:/databricks/jars/----workspace_spark_3_0--common--tags--tags-hive-2.3__hadoop-2.7_2.12_deploy.jar:/databricks/jars/----workspace_spark_3_0--common--unsafe--unsafe-hive-2.3__hadoop-2.7_2.12_deploy.jar:/databricks/jars/----workspace_spark_3_0--core--core-hive-2.3__hadoop-2.7_2.12_deploy.jar:/databricks/jars/----workspace_spark_3_0--core--libcore_generated_resources.jar:/databricks/jars/----workspace_spark_3_0--core--libcore_resources.jar:/databricks/jars/----workspace_spark_3_0--graphx--graphx-hive-2.3__hadoop-2.7_2.12_deploy.jar:/databricks/jars/----workspace_spark_3_0--launcher--launcher-hive-2.3__hadoop-2.7_2.12_deploy.jar:/databricks/jars/----workspace_spark_3_0--maven-trees--hive-2.3__hadoop-2.7--antlr--antlr--antlr__antlr__2.7.7.jar:/databricks/jars/----workspace_spark_3_0--maven-trees--hive-2.3__hadoop-2.7--com.amazonaws--amazon-kinesis-client--com.amazonaws__amazon-kinesis-client__1.12.0.jar:/databricks/jars/----workspace_spark_3_0--maven-trees--hive-2.3__hadoop-2.7--com.amazonaws--aws-java-sdk-autoscaling--com.amazonaws__aws-java-sdk-autoscaling__1.11.655.jar:/databricks/jars/----workspace_spark_3_0--maven-trees--hive-2.3__hadoop-2.7--com.amazonaws--aws-java-sdk-cloudformation--com.amazonaws__aws-java-sdk-cloudformation__1.11.655.jar:/databricks/jars/----workspace_spark_3_0--maven-trees--hive-2.3__hadoop-2.7--com.amazonaws--aws-java-sdk-cloudfront--com.amazonaws__aws-java-sdk-cloudfront__1.11.655.jar:/databricks/jars/----workspace_spark_3_0--maven-trees--hive-2.3__hadoop-2.7--com.amazonaws--aws-java-sdk-cloudhsm--com.amazonaws__aws-java-sdk-cloudhsm__1.11.655.jar:/databricks/jars/----workspace_spark_3_0--maven-trees--hive-2.3__hadoop-2.7--com.amazonaws--aws-java-sdk-cloudsearch--com.amazonaws__aws-java-sdk-cloudsearch__1.11.655.jar:/databricks/jars/----workspace_spark_3_0--maven-trees--hive-2.3__hadoop-2.7--com.amazonaws--aws-java-sdk-cloudtrail--com.amazonaws__aws-java-sdk-cloudtrail__1.11.655.jar:/databricks/jars/----workspace_spark_3_0--maven-trees--hive-2.3__hadoop-2.7--com.amazonaws--aws-java-sdk-cloudwatch--com.amazonaws__aws-java-sdk-cloudwatch__1.11.655.jar:/databricks/jars/----workspace_spark_3_0--maven-trees--hive-2.3__hadoop-2.7--com.amazonaws--aws-java-sdk-cloudwatchmetrics--com.amazonaws__aws-java-sdk-cloudwatchmetrics__1.11.655.jar:/databricks/jars/----workspace_spark_3_0--maven-trees--hive-2.3__hadoop-2.7--com.amazonaws--aws-java-sdk-codedeploy--com.amazonaws__aws-java-sdk-codedeploy__1.11.655.jar:/databricks/jars/----workspace_spark_3_0--maven-trees--hive-2.3__hadoop-2.7--com.amazonaws--aws-java-sdk-cognitoidentity--com.amazonaws__aws-java-sdk-cognitoidentity__1.11.655.jar:/databricks/jars/----workspace_spark_3_0--maven-trees--hive-2.3__hadoop-2.7--com.amazonaws--aws-java-sdk-cognitosync--com.amazonaws__aws-java-sdk-cognitosync__1.11.655.jar:/databricks/jars/----workspace_spark_3_0--maven-trees--hive-2.3__hadoop-2.7--com.amazonaws--aws-java-sdk-config--com.amazonaws__aws-java-sdk-config__1.11.655.jar:/databricks/jars/----workspace_spark_3_0--maven-trees--hive-2.3__hadoop-2.7--com.amazonaws--aws-java-sdk-core--com.amazonaws__aws-java-sdk-core__1.11.655.jar:/databricks/jars/----workspace_spark_3_0--maven-trees--hive-2.3__hadoop-2.7--com.amazonaws--aws-java-sdk-datapipeline--com.amazonaws__aws-java-sdk-datapipeline__1.11.655.jar:/databricks/jars/----workspace_spark_3_0--maven-trees--hive-2.3__hadoop-2.7--com.amazonaws--aws-java-sdk-directconnect--com.amazonaws__aws-java-sdk-directconnect__1.11.655.jar:/databricks/jars/----workspace_spark_3_0--maven-trees--hive-2.3__hadoop-2.7--com.amazonaws--aws-java-sdk-directory--com.amazonaws__aws-java-sdk-directory__1.11.655.jar:/databricks/jars/----workspace_spark_3_0--maven-trees--hive-2.3__hadoop-2.7--com.amazonaws--aws-java-sdk-dynamodb--com.amazonaws__aws-java-sdk-dynamodb__1.11.655.jar:/databricks/jars/----workspace_spark_3_0--maven-trees--hive-2.3__hadoop-2.7--com.amazonaws--aws-java-sdk-ec2--com.amazonaws__aws-java-sdk-ec2__1.11.655.jar:/databricks/jars/----workspace_spark_3_0--maven-trees--hive-2.3__hadoop-2.7--com.amazonaws--aws-java-sdk-ecs--com.amazonaws__aws-java-sdk-ecs__1.11.655.jar:/databricks/jars/----workspace_spark_3_0--maven-trees--hive-2.3__hadoop-2.7--com.amazonaws--aws-java-sdk-efs--com.amazonaws__aws-java-sdk-efs__1.11.655.jar:/databricks/jars/----workspace_spark_3_0--maven-trees--hive-2.3__hadoop-2.7--com.amazonaws--aws-java-sdk-elasticache--com.amazonaws__aws-java-sdk-elasticache__1.11.655.jar:/databricks/jars/----workspace_spark_3_0--maven-trees--hive-2.3__hadoop-2.7--com.amazonaws--aws-java-sdk-elasticbeanstalk--com.amazonaws__aws-java-sdk-elasticbeanstalk__1.11.655.jar:/databricks/jars/----workspace_spark_3_0--maven-trees--hive-2.3__hadoop-2.7--com.amazonaws--aws-java-sdk-elasticloadbalancing--com.amazonaws__aws-java-sdk-elasticloadbalancing__1.11.655.jar:/databricks/jars/----workspace_spark_3_0--maven-trees--hive-2.3__hadoop-2.7--com.amazonaws--aws-java-sdk-elastictranscoder--com.amazonaws__aws-java-sdk-elastictranscoder__1.11.655.jar:/databricks/jars/----workspace_spark_3_0--maven-trees--hive-2.3__hadoop-2.7--com.amazonaws--aws-java-sdk-emr--com.amazonaws__aws-java-sdk-emr__1.11.655.jar:/databricks/jars/----workspace_spark_3_0--maven-trees--hive-2.3__hadoop-2.7--com.amazonaws--aws-java-sdk-glacier--com.amazonaws__aws-java-sdk-glacier__1.11.655.jar:/databricks/jars/----workspace_spark_3_0--maven-trees--hive-2.3__hadoop-2.7--com.amazonaws--aws-java-sdk-iam--com.amazonaws__aws-java-sdk-iam__1.11.655.jar:/databricks/jars/----workspace_spark_3_0--maven-trees--hive-2.3__hadoop-2.7--com.amazonaws--aws-java-sdk-importexport--com.amazonaws__aws-java-sdk-importexport__1.11.655.jar:/databricks/jars/----workspace_spark_3_0--maven-trees--hive-2.3__hadoop-2.7--com.amazonaws--aws-java-sdk-kinesis--com.amazonaws__aws-java-sdk-kinesis__1.11.655.jar:/databricks/jars/----workspace_spark_3_0--maven-trees--hive-2.3__hadoop-2.7--com.amazonaws--aws-java-sdk-kms--com.amazonaws__aws-java-sdk-kms__1.11.655.jar:/databricks/jars/----workspace_spark_3_0--maven-trees--hive-2.3__hadoop-2.7--com.amazonaws--aws-java-sdk-lambda--com.amazonaws__aws-java-sdk-lambda__1.11.655.jar:/databricks/jars/----workspace_spark_3_0--maven-trees--hive-2.3__hadoop-2.7--com.amazonaws--aws-java-sdk-logs--com.amazonaws__aws-java-sdk-logs__1.11.655.jar:/databricks/jars/----workspace_spark_3_0--maven-trees--hive-2.3__hadoop-2.7--com.amazonaws--aws-java-sdk-machinelearning--com.amazonaws__aws-java-sdk-machinelearning__1.11.655.jar:/databricks/jars/----workspace_spark_3_0--maven-trees--hive-2.3__hadoop-2.7--com.amazonaws--aws-java-sdk-opsworks--com.amazonaws__aws-java-sdk-opsworks__1.11.655.jar:/databricks/jars/----workspace_spark_3_0--maven-trees--hive-2.3__hadoop-2.7--com.amazonaws--aws-java-sdk-rds--com.amazonaws__aws-java-sdk-rds__1.11.655.jar:/databricks/jars/----workspace_spark_3_0--maven-trees--hive-2.3__hadoop-2.7--com.amazonaws--aws-java-sdk-redshift--com.amazonaws__aws-java-sdk-redshift__1.11.655.jar:/databricks/jars/----workspace_spark_3_0--maven-trees--hive-2.3__hadoop-2.7--com.amazonaws--aws-java-sdk-route53--com.amazonaws__aws-java-sdk-route53__1.11.655.jar:/databricks/jars/----workspace_spark_3_0--maven-trees--hive-2.3__hadoop-2.7--com.amazonaws--aws-java-sdk-s3--com.amazonaws__aws-java-sdk-s3__1.11.655.jar:/databricks/jars/----workspace_spark_3_0--maven-trees--hive-2.3__hadoop-2.7--com.amazonaws--aws-java-sdk-ses--com.amazonaws__aws-java-sdk-ses__1.11.655.jar:/databricks/jars/----workspace_spark_3_0--maven-trees--hive-2.3__hadoop-2.7--com.amazonaws--aws-java-sdk-simpledb--com.amazonaws__aws-java-sdk-simpledb__1.11.655.jar:/databricks/jars/----workspace_spark_3_0--maven-trees--hive-2.3__hadoop-2.7--com.amazonaws--aws-java-sdk-simpleworkflow--com.amazonaws__aws-java-sdk-simpleworkflow__1.11.655.jar:/databricks/jars/----workspace_spark_3_0--maven-trees--hive-2.3__hadoop-2.7--com.amazonaws--aws-java-sdk-sns--com.amazonaws__aws-java-sdk-sns__1.11.655.jar:/databricks/jars/----workspace_spark_3_0--maven-trees--hive-2.3__hadoop-2.7--com.amazonaws--aws-java-sdk-sqs--com.amazonaws__aws-java-sdk-sqs__1.11.655.jar:/databricks/jars/----workspace_spark_3_0--maven-trees--hive-2.3__hadoop-2.7--com.amazonaws--aws-java-sdk-ssm--com.amazonaws__aws-java-sdk-ssm__1.11.655.jar:/databricks/jars/----workspace_spark_3_0--maven-trees--hive-2.3__hadoop-2.7--com.amazonaws--aws-java-sdk-storagegateway--com.amazonaws__aws-java-sdk-storagegateway__1.11.655.jar:/databricks/jars/----workspace_spark_3_0--maven-trees--hive-2.3__hadoop-2.7--com.amazonaws--aws-java-sdk-sts--com.amazonaws__aws-java-sdk-sts__1.11.655.jar:/databricks/jars/----workspace_spark_3_0--maven-trees--hive-2.3__hadoop-2.7--com.amazonaws--aws-java-sdk-support--com.amazonaws__aws-java-sdk-support__1.11.655.jar:/databricks/jars/----workspace_spark_3_0--maven-trees--hive-2.3__hadoop-2.7--com.amazonaws--aws-java-sdk-swf-libraries--com.amazonaws__aws-java-sdk-swf-libraries__1.11.22.jar:/databricks/jars/----workspace_spark_3_0--maven-trees--hive-2.3__hadoop-2.7--com.amazonaws--aws-java-sdk-workspaces--com.amazonaws__aws-java-sdk-workspaces__1.11.655.jar:/databricks/jars/----workspace_spark_3_0--maven-trees--hive-2.3__hadoop-2.7--com.amazonaws--jmespath-java--com.amazonaws__jmespath-java__1.11.655.jar:/databricks/jars/----workspace_spark_3_0--maven-trees--hive-2.3__hadoop-2.7--com.chuusai--shapeless_2.12--com.chuusai__shapeless_2.12__2.3.3.jar:/databricks/jars/----workspace_spark_3_0--maven-trees--hive-2.3__hadoop-2.7--com.clearspring.analytics--stream--com.clearspring.analytics__stream__2.9.6.jar:/databricks/jars/----workspace_spark_3_0--maven-trees--hive-2.3__hadoop-2.7--com.databricks--Rserve--com.databricks__Rserve__1.8-3.jar:/databricks/jars/----workspace_spark_3_0--maven-trees--hive-2.3__hadoop-2.7--com.databricks.scalapb--compilerplugin_2.12--com.databricks.scalapb__compilerplugin_2.12__0.4.15-10.jar:/databricks/jars/----workspace_spark_3_0--maven-trees--hive-2.3__hadoop-2.7--com.databricks.scalapb--scalapb-runtime_2.12--com.databricks.scalapb__scalapb-runtime_2.12__0.4.15-10.jar:/databricks/jars/----workspace_spark_3_0--maven-trees--hive-2.3__hadoop-2.7--com.esotericsoftware--kryo-shaded--com.esotericsoftware__kryo-shaded__4.0.2.jar:/databricks/jars/----workspace_spark_3_0--maven-trees--hive-2.3__hadoop-2.7--com.esotericsoftware--minlog--com.esotericsoftware__minlog__1.3.0.jar:/databricks/jars/----workspace_spark_3_0--maven-trees--hive-2.3__hadoop-2.7--com.fasterxml--classmate--com.fasterxml__classmate__1.3.4.jar:/databricks/jars/----workspace_spark_3_0--maven-trees--hive-2.3__hadoop-2.7--com.fasterxml.jackson.core--jackson-annotations--com.fasterxml.jackson.core__jackson-annotations__2.10.0.jar:/databricks/jars/----workspace_spark_3_0--maven-trees--hive-2.3__hadoop-2.7--com.fasterxml.jackson.core--jackson-core--com.fasterxml.jackson.core__jackson-core__2.10.0.jar:/databricks/jars/----workspace_spark_3_0--maven-trees--hive-2.3__hadoop-2.7--com.fasterxml.jackson.core--jackson-databind--com.fasterxml.jackson.core__jackson-databind__2.10.0.jar:/databricks/jars/----workspace_spark_3_0--maven-trees--hive-2.3__hadoop-2.7--com.fasterxml.jackson.dataformat--jackson-dataformat-cbor--com.fasterxml.jackson.dataformat__jackson-dataformat-cbor__2.10.0.jar:/databricks/jars/----workspace_spark_3_0--maven-trees--hive-2.3__hadoop-2.7--com.fasterxml.jackson.datatype--jackson-datatype-joda--com.fasterxml.jackson.datatype__jackson-datatype-joda__2.10.0.jar:/databricks/jars/----workspace_spark_3_0--maven-trees--hive-2.3__hadoop-2.7--com.fasterxml.jackson.module--jackson-module-paranamer--com.fasterxml.jackson.module__jackson-module-paranamer__2.10.0.jar:/databricks/jars/----workspace_spark_3_0--maven-trees--hive-2.3__hadoop-2.7--com.fasterxml.jackson.module--jackson-module-scala_2.12--com.fasterxml.jackson.module__jackson-module-scala_2.12__2.10.0.jar:/databricks/jars/----workspace_spark_3_0--maven-trees--hive-2.3__hadoop-2.7--com.github.ben-manes.caffeine--caffeine--com.github.ben-manes.caffeine__caffeine__2.3.4.jar:/databricks/jars/----workspace_spark_3_0--maven-trees--hive-2.3__hadoop-2.7--com.github.fommil--jniloader--com.github.fommil__jniloader__1.1.jar:/databricks/jars/----workspace_spark_3_0--maven-trees--hive-2.3__hadoop-2.7--com.github.fommil.netlib--core--com.github.fommil.netlib__core__1.1.2.jar:/databricks/jars/----workspace_spark_3_0--maven-trees--hive-2.3__hadoop-2.7--com.github.fommil.netlib--native_ref-java--com.github.fommil.netlib__native_ref-java__1.1.jar:/databricks/jars/----workspace_spark_3_0--maven-trees--hive-2.3__hadoop-2.7--com.github.fommil.netlib--native_ref-java-natives--com.github.fommil.netlib__native_ref-java-natives__1.1.jar:/databricks/jars/----workspace_spark_3_0--maven-trees--hive-2.3__hadoop-2.7--com.github.fommil.netlib--native_system-java--com.github.fommil.netlib__native_system-java__1.1.jar:/databricks/jars/----workspace_spark_3_0--maven-trees--hive-2.3__hadoop-2.7--com.github.fommil.netlib--native_system-java-natives--com.github.fommil.netlib__native_system-java-natives__1.1.jar:/databricks/jars/----workspace_spark_3_0--maven-trees--hive-2.3__hadoop-2.7--com.github.fommil.netlib--netlib-native_ref-linux-x86_64-natives--com.github.fommil.netlib__netlib-native_ref-linux-x86_64-natives__1.1.jar:/databricks/jars/----workspace_spark_3_0--maven-trees--hive-2.3__hadoop-2.7--com.github.fommil.netlib--netlib-native_system-linux-x86_64-natives--com.github.fommil.netlib__netlib-native_system-linux-x86_64-natives__1.1.jar:/databricks/jars/----workspace_spark_3_0--maven-trees--hive-2.3__hadoop-2.7--com.github.joshelser--dropwizard-metrics-hadoop-metrics2-reporter--com.github.joshelser__dropwizard-metrics-hadoop-metrics2-reporter__0.1.2.jar:/databricks/jars/----workspace_spark_3_0--maven-trees--hive-2.3__hadoop-2.7--com.github.luben--zstd-jni--com.github.luben__zstd-jni__1.4.4-3.jar:/databricks/jars/----workspace_spark_3_0--maven-trees--hive-2.3__hadoop-2.7--com.github.wendykierp--JTransforms--com.github.wendykierp__JTransforms__3.1.jar:/databricks/jars/----workspace_spark_3_0--maven-trees--hive-2.3__hadoop-2.7--com.google.code.findbugs--jsr305--com.google.code.findbugs__jsr305__3.0.0.jar:/databricks/jars/----workspace_spark_3_0--maven-trees--hive-2.3__hadoop-2.7--com.google.code.gson--gson--com.google.code.gson__gson__2.2.4.jar:/databricks/jars/----workspace_spark_3_0--maven-trees--hive-2.3__hadoop-2.7--com.google.flatbuffers--flatbuffers-java--com.google.flatbuffers__flatbuffers-java__1.9.0.jar:/databricks/jars/----workspace_spark_3_0--maven-trees--hive-2.3__hadoop-2.7--com.google.guava--guava--com.google.guava__guava__15.0.jar:/databricks/jars/----workspace_spark_3_0--maven-trees--hive-2.3__hadoop-2.7--com.google.protobuf--protobuf-java--com.google.protobuf__protobuf-java__2.6.1.jar:/databricks/jars/----workspace_spark_3_0--maven-trees--hive-2.3__hadoop-2.7--com.h2database--h2--com.h2database__h2__1.4.195.jar:/databricks/jars/----workspace_spark_3_0--maven-trees--hive-2.3__hadoop-2.7--com.helger--profiler--com.helger__profiler__1.1.1.jar:/databricks/jars/----workspace_spark_3_0--maven-trees--hive-2.3__hadoop-2.7--com.jcraft--jsch--com.jcraft__jsch__0.1.50.jar:/databricks/jars/----workspace_spark_3_0--maven-trees--hive-2.3__hadoop-2.7--com.jolbox--bonecp--com.jolbox__bonecp__0.8.0.RELEASE.jar:/databricks/jars/----workspace_spark_3_0--maven-trees--hive-2.3__hadoop-2.7--com.lihaoyi--sourcecode_2.12--com.lihaoyi__sourcecode_2.12__0.1.9.jar:/databricks/jars/----workspace_spark_3_0--maven-trees--hive-2.3__hadoop-2.7--com.microsoft.azure--azure-data-lake-store-sdk--com.microsoft.azure__azure-data-lake-store-sdk__2.2.8.jar:/databricks/jars/----workspace_spark_3_0--maven-trees--hive-2.3__hadoop-2.7--com.microsoft.sqlserver--mssql-jdbc--com.microsoft.sqlserver__mssql-jdbc__8.2.1.jre8.jar:/databricks/jars/----workspace_spark_3_0--maven-trees--hive-2.3__hadoop-2.7--commons-beanutils--commons-beanutils--commons-beanutils__commons-beanutils__1.9.4.jar:/databricks/jars/----workspace_spark_3_0--maven-trees--hive-2.3__hadoop-2.7--commons-cli--commons-cli--commons-cli__commons-cli__1.2.jar:/databricks/jars/----workspace_spark_3_0--maven-trees--hive-2.3__hadoop-2.7--commons-codec--commons-codec--commons-codec__commons-codec__1.10.jar:/databricks/jars/----workspace_spark_3_0--maven-trees--hive-2.3__hadoop-2.7--commons-collections--commons-collections--commons-collections__commons-collections__3.2.2.jar:/databricks/jars/----workspace_spark_3_0--maven-trees--hive-2.3__hadoop-2.7--commons-configuration--commons-configuration--commons-configuration__commons-configuration__1.6.jar:/databricks/jars/----workspace_spark_3_0--maven-trees--hive-2.3__hadoop-2.7--commons-dbcp--commons-dbcp--commons-dbcp__commons-dbcp__1.4.jar:/databricks/jars/----workspace_spark_3_0--maven-trees--hive-2.3__hadoop-2.7--commons-digester--commons-digester--commons-digester__commons-digester__1.8.jar:/databricks/jars/----workspace_spark_3_0--maven-trees--hive-2.3__hadoop-2.7--commons-fileupload--commons-fileupload--commons-fileupload__commons-fileupload__1.3.3.jar:/databricks/jars/----workspace_spark_3_0--maven-trees--hive-2.3__hadoop-2.7--commons-httpclient--commons-httpclient--commons-httpclient__commons-httpclient__3.1.jar:/databricks/jars/----workspace_spark_3_0--maven-trees--hive-2.3__hadoop-2.7--commons-io--commons-io--commons-io__commons-io__2.4.jar:/databricks/jars/----workspace_spark_3_0--maven-trees--hive-2.3__hadoop-2.7--commons-lang--commons-lang--commons-lang__commons-lang__2.6.jar:/databricks/jars/----workspace_spark_3_0--maven-trees--hive-2.3__hadoop-2.7--commons-logging--commons-logging--commons-logging__commons-logging__1.1.3.jar:/databricks/jars/----workspace_spark_3_0--maven-trees--hive-2.3__hadoop-2.7--commons-net--commons-net--commons-net__commons-net__3.1.jar:/databricks/jars/----workspace_spark_3_0--maven-trees--hive-2.3__hadoop-2.7--commons-pool--commons-pool--commons-pool__commons-pool__1.5.4.jar:/databricks/jars/----workspace_spark_3_0--maven-trees--hive-2.3__hadoop-2.7--com.ning--compress-lzf--com.ning__compress-lzf__1.0.3.jar:/databricks/jars/----workspace_spark_3_0--maven-trees--hive-2.3__hadoop-2.7--com.sun.mail--javax.mail--com.sun.mail__javax.mail__1.5.2.jar:/databricks/jars/----workspace_spark_3_0--maven-trees--hive-2.3__hadoop-2.7--com.tdunning--json--com.tdunning__json__1.8.jar:/databricks/jars/----workspace_spark_3_0--maven-trees--hive-2.3__hadoop-2.7--com.thoughtworks.paranamer--paranamer--com.thoughtworks.paranamer__paranamer__2.8.jar:/databricks/jars/----workspace_spark_3_0--maven-trees--hive-2.3__hadoop-2.7--com.trueaccord.lenses--lenses_2.12--com.trueaccord.lenses__lenses_2.12__0.4.12.jar:/databricks/jars/----workspace_spark_3_0--maven-trees--hive-2.3__hadoop-2.7--com.twitter--chill_2.12--com.twitter__chill_2.12__0.9.5.jar:/databricks/jars/----workspace_spark_3_0--maven-trees--hive-2.3__hadoop-2.7--com.twitter--chill-java--com.twitter__chill-java__0.9.5.jar:/databricks/jars/----workspace_spark_3_0--maven-trees--hive-2.3__hadoop-2.7--com.twitter--util-app_2.12--com.twitter__util-app_2.12__7.1.0.jar:/databricks/jars/----workspace_spark_3_0--maven-trees--hive-2.3__hadoop-2.7--com.twitter--util-core_2.12--com.twitter__util-core_2.12__7.1.0.jar:/databricks/jars/----workspace_spark_3_0--maven-trees--hive-2.3__hadoop-2.7--com.twitter--util-function_2.12--com.twitter__util-function_2.12__7.1.0.jar:/databricks/jars/----workspace_spark_3_0--maven-trees--hive-2.3__hadoop-2.7--com.twitter--util-jvm_2.12--com.twitter__util-jvm_2.12__7.1.0.jar:/databricks/jars/----workspace_spark_3_0--ma
*** WARNING: skipped 122159 bytes of output ***
container=lxc
USER=root
PYTHONHASHSEED=0
MLFLOW_PYTHON_EXECUTABLE=/databricks/python/bin/python
PWD=/databricks/driver
R_LIBS=/databricks/spark/R/lib
HOME=/root
DB_HOME=/databricks
SPARK_LOCAL_DIRS=/local_disk0
SUDO_USER=root
PYSPARK_GATEWAY_PORT=40242
DATABRICKS_ROOT_VIRTUALENV_ENV=/databricks/python3
ENABLE_IPTABLES=false
SPARK_LOCAL_IP=10.149.224.176
SPARK_SCALA_VERSION=2.12
SPARK_HOME=/databricks/spark
CLUSTER_DB_HOME=/databricks
SUDO_UID=0
SPARK_WORKER_MEMORY=3146m
MAIL=/var/mail/root
TERM=unknown
SHELL=/bin/bash
MPLBACKEND=AGG
DATABRICKS_RUNTIME_VERSION=7.2
SPARK_ENV_LOADED=1
SHLVL=4
PYTHONPATH=/databricks/spark/python:/databricks/spark/python/lib/py4j-0.10.9-src.zip:/databricks/jars/spark--driver--driver-spark_3.0_2.12_deploy.jar:/databricks/spark/python
SCALA_VERSION=2.10
PYSPARK_GATEWAY_SECRET=cbfa4e3656b7584c09eb35b9ceeaf9b9369fac897eb3569e0ccfc719dd762523
LOGNAME=root
MLFLOW_TRACKING_URI=databricks
SPARK_CONF_DIR=/databricks/spark/conf
PYSPARK_PYTHON=/databricks/python/bin/python
KOALAS_USAGE_LOGGER=pyspark.databricks.koalas.usage_logger
PATH=/databricks/python3/bin:/usr/local/sbin:/usr/local/bin:/usr/sbin:/usr/bin:/sbin:/bin:/snap/bin
DRIVER_PID_FILE=/tmp/driver-daemon.pid
PIP_NO_INPUT=1
tar zxvf sou.tgz
sou/
sou/18101205.txt
sou/19390104.txt
sou/18361205.txt
sou/18701205.txt
sou/17911025.txt
sou/18511202.txt
sou/18661203.txt
sou/18751207.txt
sou/18731201.txt
sou/18861206.txt
sou/19470106.txt
sou/18721202.txt
sou/19690114.txt
sou/19980127.txt
sou/18241207.txt
sou/19440111.txt
sou/18261205.txt
sou/18501202.txt
sou/18081108.txt
sou/18421206.txt
sou/19870127.txt
sou/19490105.txt
sou/19520109.txt
sou/19131202.txt
sou/18811206.txt
sou/19460121.txt
sou/18201114.txt
sou/18021215.txt
sou/18371205.txt
sou/18491204.txt
sou/19350104.txt
sou/19610112.txt
sou/19360103.txt
sou/18881203.txt
sou/18671203.txt
sou/19171204.txt
sou/20090224.txt
sou/18921206.txt
sou/19271206.txt
sou/18691206.txt
sou/19720120.txt
sou/20100127.txt
sou/17951208.txt
sou/19071203.txt
sou/18651204.txt
sou/18971206.txt
sou/19800121.txt
sou/19251208.txt
sou/19700122.txt
sou/18231202.txt
sou/20010227.txt
sou/17931203.txt
sou/18441203.txt
sou/19920128.txt
sou/18471207.txt
sou/18951207.txt
sou/19081208.txt
sou/18251206.txt
sou/19830125.txt
sou/18411207.txt
sou/18771203.txt
sou/19590109.txt
sou/18351207.txt
sou/18601203.txt
sou/18291208.txt
sou/17991203.txt
sou/19610130.txt
sou/20060131.txt
sou/19580109.txt
sou/18061202.txt
sou/18831204.txt
sou/19780119.txt
sou/19061203.txt
sou/17981208.txt
sou/19650104.txt
sou/19161205.txt
sou/19740130.txt
sou/19940125.txt
sou/17901208.txt
sou/19001203.txt
sou/18181116.txt
sou/18871206.txt
sou/19051205.txt
sou/19450106.txt
sou/19231206.txt
sou/19970204.txt
sou/19640108.txt
sou/19261207.txt
sou/20140128.txt
sou/19770112.txt
sou/20030128.txt
sou/19840125.txt
sou/18401205.txt
sou/19430107.txt
sou/17971122.txt
sou/19311208.txt
sou/18091129.txt
sou/18781202.txt
sou/18521206.txt
sou/18851208.txt
sou/19570110.txt
sou/19750115.txt
sou/19370106.txt
sou/19281204.txt
sou/17921106.txt
sou/19620111.txt
sou/18121104.txt
sou/19221208.txt
sou/18531205.txt
sou/19550106.txt
sou/18581206.txt
sou/19540107.txt
sou/19880125.txt
sou/19211206.txt
sou/18281202.txt
sou/19730202.txt
sou/18681209.txt
sou/20080128.txt
sou/18841201.txt
sou/18551231.txt
sou/19041206.txt
sou/18011208.txt
sou/20040120.txt
sou/18131207.txt
sou/18991205.txt
sou/18031017.txt
sou/19530107.txt
sou/19201207.txt
sou/19710122.txt
sou/18791201.txt
sou/19480107.txt
sou/18271204.txt
sou/17900108.txt
sou/19930217.txt
sou/18911209.txt
sou/18641206.txt
sou/19021202.txt
sou/18891203.txt
sou/18961204.txt
sou/19530202.txt
sou/20120124.txt
sou/18481205.txt
sou/19860204.txt
sou/19400103.txt
sou/19340103.txt
sou/19630114.txt
sou/19850206.txt
sou/18051203.txt
sou/19510108.txt
sou/19890209.txt
sou/19560105.txt
sou/19810116.txt
sou/19380103.txt
sou/20010920.txt
sou/20000127.txt
sou/19241203.txt
sou/19091207.txt
sou/19011203.txt
sou/18001111.txt
sou/18111105.txt
sou/19820126.txt
sou/19500104.txt
sou/18171212.txt
sou/19101206.txt
sou/18461208.txt
sou/19600107.txt
sou/18451202.txt
sou/20160112.txt
sou/18311206.txt
sou/19960123.txt
sou/19680117.txt
sou/18161203.txt
sou/19151207.txt
sou/18151205.txt
sou/18621201.txt
sou/19301202.txt
sou/19191202.txt
sou/19790125.txt
sou/19670110.txt
sou/18901201.txt
sou/18071027.txt
sou/18140920.txt
sou/20170228.txt
sou/19990119.txt
sou/20020129.txt
sou/19760119.txt
sou/18801206.txt
sou/18741207.txt
sou/19121203.txt
sou/18941202.txt
sou/20110125.txt
sou/18631208.txt
sou/18041108.txt
sou/18561202.txt
sou/19410106.txt
sou/20070123.txt
sou/18211203.txt
sou/19181202.txt
sou/20150120.txt
sou/18541204.txt
sou/18931203.txt
sou/18591219.txt
sou/18571208.txt
sou/19141208.txt
sou/19291203.txt
sou/17961207.txt
sou/18391202.txt
sou/18761205.txt
sou/18341201.txt
sou/18821204.txt
sou/18221203.txt
sou/18981205.txt
sou/19900131.txt
sou/19031207.txt
sou/19111205.txt
sou/18301206.txt
sou/19950124.txt
sou/18431206.txt
sou/20130212.txt
sou/17941119.txt
sou/19660112.txt
sou/18321204.txt
sou/19420106.txt
sou/18381203.txt
sou/19910129.txt
sou/18711204.txt
sou/18331203.txt
sou/19321206.txt
sou/20050202.txt
sou/18611203.txt
sou/18191207.txt
cd sou && ls
17900108.txt
17901208.txt
17911025.txt
17921106.txt
17931203.txt
17941119.txt
17951208.txt
17961207.txt
17971122.txt
17981208.txt
17991203.txt
18001111.txt
18011208.txt
18021215.txt
18031017.txt
18041108.txt
18051203.txt
18061202.txt
18071027.txt
18081108.txt
18091129.txt
18101205.txt
18111105.txt
18121104.txt
18131207.txt
18140920.txt
18151205.txt
18161203.txt
18171212.txt
18181116.txt
18191207.txt
18201114.txt
18211203.txt
18221203.txt
18231202.txt
18241207.txt
18251206.txt
18261205.txt
18271204.txt
18281202.txt
18291208.txt
18301206.txt
18311206.txt
18321204.txt
18331203.txt
18341201.txt
18351207.txt
18361205.txt
18371205.txt
18381203.txt
18391202.txt
18401205.txt
18411207.txt
18421206.txt
18431206.txt
18441203.txt
18451202.txt
18461208.txt
18471207.txt
18481205.txt
18491204.txt
18501202.txt
18511202.txt
18521206.txt
18531205.txt
18541204.txt
18551231.txt
18561202.txt
18571208.txt
18581206.txt
18591219.txt
18601203.txt
18611203.txt
18621201.txt
18631208.txt
18641206.txt
18651204.txt
18661203.txt
18671203.txt
18681209.txt
18691206.txt
18701205.txt
18711204.txt
18721202.txt
18731201.txt
18741207.txt
18751207.txt
18761205.txt
18771203.txt
18781202.txt
18791201.txt
18801206.txt
18811206.txt
18821204.txt
18831204.txt
18841201.txt
18851208.txt
18861206.txt
18871206.txt
18881203.txt
18891203.txt
18901201.txt
18911209.txt
18921206.txt
18931203.txt
18941202.txt
18951207.txt
18961204.txt
18971206.txt
18981205.txt
18991205.txt
19001203.txt
19011203.txt
19021202.txt
19031207.txt
19041206.txt
19051205.txt
19061203.txt
19071203.txt
19081208.txt
19091207.txt
19101206.txt
19111205.txt
19121203.txt
19131202.txt
19141208.txt
19151207.txt
19161205.txt
19171204.txt
19181202.txt
19191202.txt
19201207.txt
19211206.txt
19221208.txt
19231206.txt
19241203.txt
19251208.txt
19261207.txt
19271206.txt
19281204.txt
19291203.txt
19301202.txt
19311208.txt
19321206.txt
19340103.txt
19350104.txt
19360103.txt
19370106.txt
19380103.txt
19390104.txt
19400103.txt
19410106.txt
19420106.txt
19430107.txt
19440111.txt
19450106.txt
19460121.txt
19470106.txt
19480107.txt
19490105.txt
19500104.txt
19510108.txt
19520109.txt
19530107.txt
19530202.txt
19540107.txt
19550106.txt
19560105.txt
19570110.txt
19580109.txt
19590109.txt
19600107.txt
19610112.txt
19610130.txt
19620111.txt
19630114.txt
19640108.txt
19650104.txt
19660112.txt
19670110.txt
19680117.txt
19690114.txt
19700122.txt
19710122.txt
19720120.txt
19730202.txt
19740130.txt
19750115.txt
19760119.txt
19770112.txt
19780119.txt
19790125.txt
19800121.txt
19810116.txt
19820126.txt
19830125.txt
19840125.txt
19850206.txt
19860204.txt
19870127.txt
19880125.txt
19890209.txt
19900131.txt
19910129.txt
19920128.txt
19930217.txt
19940125.txt
19950124.txt
19960123.txt
19970204.txt
19980127.txt
19990119.txt
20000127.txt
20010227.txt
20010920.txt
20020129.txt
20030128.txt
20040120.txt
20050202.txt
20060131.txt
20070123.txt
20080128.txt
20090224.txt
20100127.txt
20110125.txt
20120124.txt
20130212.txt
20140128.txt
20150120.txt
20160112.txt
20170228.txt
head sou/17900108.txt
George Washington
January 8, 1790
Fellow-Citizens of the Senate and House of Representatives:
I embrace with great satisfaction the opportunity which now presents itself of congratulating you on the present favorable prospects of our public affairs. The recent accession of the important state of North Carolina to the Constitution of the United States (of which official information has been received), the rising credit and respectability of our country, the general and increasing good will toward the government of the Union, and the concord, peace, and plenty with which we are blessed are circumstances auspicious in an eminent degree to our national prosperity.
In resuming your consultations for the general good you can not but derive encouragement from the reflection that the measures of the last session have been as satisfactory to your constituents as the novelty and difficulty of the work allowed you to hope. Still further to realize their expectations and to secure the blessings which a gracious Providence has placed within our reach will in the course of the present important session call for the cool and deliberate exertion of your patriotism, firmness, and wisdom.
Among the many interesting objects which will engage your attention that of providing for the common defense will merit particular regard. To be prepared for war is one of the most effectual means of preserving peace.
A free people ought not only to be armed, but disciplined; to which end a uniform and well-digested plan is requisite; and their safety and interest require that they should promote such manufactories as tend to render them independent of others for essential, particularly military, supplies.
The proper establishment of the troops which may be deemed indispensable will be entitled to mature consideration. In the arrangements which may be made respecting it it will be of importance to conciliate the comfortable support of the officers and soldiers with a due regard to economy.
There was reason to hope that the pacific measures adopted with regard to certain hostile tribes of Indians would have relieved the inhabitants of our southern and western frontiers from their depredations, but you will perceive from the information contained in the papers which I shall direct to be laid before you (comprehending a communication from the Commonwealth of Virginia) that we ought to be prepared to afford protection to those parts of the Union, and, if necessary, to punish aggressors.
tail sou/17900108.txt
The advancement of agriculture, commerce, and manufactures by all proper means will not, I trust, need recommendation; but I can not forbear intimating to you the expediency of giving effectual encouragement as well to the introduction of new and useful inventions from abroad as to the exertions of skill and genius in producing them at home, and of facilitating the intercourse between the distant parts of our country by a due attention to the post-office and post-roads.
Nor am I less persuaded that you will agree with me in opinion that there is nothing which can better deserve your patronage than the promotion of science and literature. Knowledge is in every country the surest basis of public happiness. In one in which the measures of government receive their impressions so immediately from the sense of the community as in ours it is proportionably essential.
To the security of a free constitution it contributes in various ways--by convincing those who are intrusted with the public administration that every valuable end of government is best answered by the enlightened confidence of the people, and by teaching the people themselves to know and to value their own rights; to discern and provide against invasions of them; to distinguish between oppression and the necessary exercise of lawful authority; between burthens proceeding from a disregard to their convenience and those resulting from the inevitable exigencies of society; to discriminate the spirit of liberty from that of licentiousness-- cherishing the first, avoiding the last--and uniting a speedy but temperate vigilance against encroachments, with an inviolable respect to the laws.
Whether this desirable object will be best promoted by affording aids to seminaries of learning already established, by the institution of a national university, or by any other expedients will be well worthy of a place in the deliberations of the legislature.
Gentlemen of the House of Representatives:
I saw with peculiar pleasure at the close of the last session the resolution entered into by you expressive of your opinion that an adequate provision for the support of the public credit is a matter of high importance to the national honor and prosperity. In this sentiment I entirely concur; and to a perfect confidence in your best endeavors to devise such a provision as will be truly with the end I add an equal reliance on the cheerful cooperation of the other branch of the legislature.
It would be superfluous to specify inducements to a measure in which the character and interests of the United States are so obviously so deeply concerned, and which has received so explicit a sanction from your declaration.
Gentlemen of the Senate and House of Representatives:
I have directed the proper officers to lay before you, respectively, such papers and estimates as regard the affairs particularly recommended to your consideration, and necessary to convey to you that information of the state of the Union which it is my duty to afford.
The welfare of our country is the great object to which our cares and efforts ought to be directed, and I shall derive great satisfaction from a cooperation with you in the pleasing though arduous task of insuring to our fellow citizens the blessings which they have a right to expect from a free, efficient, and equal government.
head sou/20150120.txt
Barack Obama
January 20, 2015
Mr. Speaker, Mr. Vice President, Members of Congress, my fellow Americans: We are 15 years into this new century. Fifteen years that dawned with terror touching our shores, that unfolded with a new generation fighting two long and costly wars, that saw a vicious recession spread across our Nation and the world. It has been and still is a hard time for many.
But tonight we turn the page. Tonight, after a breakthrough year for America, our economy is growing and creating jobs at the fastest pace since 1999. Our unemployment rate is now lower than it was before the financial crisis. More of our kids are graduating than ever before. More of our people are insured than ever before. And we are as free from the grip of foreign oil as we've been in almost 30 years.
Tonight, for the first time since 9/11, our combat mission in Afghanistan is over. Six years ago, nearly 180,000 American troops served in Iraq and Afghanistan. Today, fewer than 15,000 remain. And we salute the courage and sacrifice of every man and woman in this 9/11 generation who has served to keep us safe. We are humbled and grateful for your service.
America, for all that we have endured, for all the grit and hard work required to come back, for all the tasks that lie ahead, know this: The shadow of crisis has passed, and the State of the Union is strong.
At this momentwith a growing economy, shrinking deficits, bustling industry, booming energy productionwe have risen from recession freer to write our own future than any other nation on Earth. It's now up to us to choose who we want to be over the next 15 years and for decades to come.
Will we accept an economy where only a few of us do spectacularly well? Or will we commit ourselves to an economy that generates rising incomes and chances for everyone who makes the effort?
Will we approach the world fearful and reactive, dragged into costly conflicts that strain our military and set back our standing? Or will we lead wisely, using all elements of our power to defeat new threats and protect our planet?
tail sou/20150120.txt
We may have different takes on the events of Ferguson and New York. But surely we can understand a father who fears his son can't walk home without being harassed. And surely we can understand the wife who won't rest until the police officer she married walks through the front door at the end of his shift. And surely we can agree that it's a good thing that for the first time in 40 years, the crime rate and the incarceration rate have come down together, and use that as a starting point for Democrats and Republicans, community leaders and law enforcement, to reform America's criminal justice system so that it protects and serves all of us.
That's a better politics. That's how we start rebuilding trust. That's how we move this country forward. That's what the American people want. And that's what they deserve.
I have no more campaigns to run.
My only agenda
I know because I won both of them. My only agenda for the next 2 years is the same as the one I've had since the day I swore an oath on the steps of this Capitol: to do what I believe is best for America. If you share the broad vision I outlined tonight, I ask you to join me in the work at hand. If you disagree with parts of it, I hope you'll at least work with me where you do agree. And I commit to every Republican here tonight that I will not only seek out your ideas, I will seek to work with you to make this country stronger.
Because I want this Chamber, I want this city to reflect the truth: that for all our blind spots and shortcomings, we are a people with the strength and generosity of spirit to bridge divides, to unite in common effort, to help our neighbors, whether down the street or on the other side of the world.
I want our actions to tell every child in every neighborhood, your life matters, and we are committed to improving your life chances, as committed as we are to working on behalf of our own kids. I want future generations to know that we are a people who see our differences as a great gift, that we're a people who value the dignity and worth of every citizen: man and woman, young and old, Black and White, Latino, Asian, immigrant, Native American, gay, straight, Americans with mental illness or physical disability. Everybody matters. I want them to grow up in a country that shows the world what we still know to be true: that we are still more than a collection of red States and blue States, that we are the United States of America.
I want them to grow up in a country where a young mom can sit down and write a letter to her President with a story that sums up these past 6 years: "It's amazing what you can bounce back from when you have to. . . . We are a strong, tight-knit family who's made it through some very, very hard times."
My fellow Americans, we too are a strong, tight-knit family. We too have made it through some hard times. Fifteen years into this new century, we have picked ourselves up, dusted ourselves off, and begun again the work of remaking America. We have laid a new foundation. A brighter future is ours to write. Let's begin this new chapter together, and let's start the work right now.
Thank you. God bless you. God bless this country we love. Thank you.
head sou/20170228.txt
Donald J. Trump
February 28, 2017
Thank you very much. Mr. Speaker, Mr. Vice President, members of Congress, the first lady of the United States ...
... and citizens of America, tonight, as we mark the conclusion of our celebration of Black History Month, we are reminded of our nation's path toward civil rights and the work that still remains to be done.
Recent threats ...
Recent threats targeting Jewish community centers and vandalism of Jewish cemeteries, as well as last week's shooting in Kansas City, remind us that while we may be a nation divided on policies, we are a country that stands united in condemning hate and evil in all of its very ugly forms.
Each American generation passes the torch of truth, liberty and justice, in an unbroken chain all the way down to the present. That torch is now in our hands. And we will use it to light up the world.
I am here tonight to deliver a message of unity and strength, and it is a message deeply delivered from my heart. A new chapter ...
... of American greatness is now beginning. A new national pride is sweeping across our nation. And a new surge of optimism is placing impossible dreams firmly within our grasp. What we are witnessing today is the renewal of the American spirit. Our allies will find that America is once again ready to lead.
tail sou/20170228.txt
America is willing to find new friends, and to forge new partnerships, where shared interests align. We want harmony and stability, not war and conflict. We want peace, wherever peace can be found. America is friends today with former enemies. Some of our closest allies, decades ago, fought on the opposite side of these terrible, terrible wars. This history should give us all faith in the possibilities for a better world.
Hopefully, the 250th year for America will see a world that is more peaceful, more just, and more free.
On our 100th anniversary in 1876, citizens from across our nation came to Philadelphia to celebrate America's centennial. At that celebration, the country's builders and artists and inventors showed off their wonderful creations. Alexander Graham Bell displayed his telephone for the first time. Remington unveiled the first typewriter. An early attempt was made at electric light. Thomas Edison showed an automatic telegraph and an electric pen. Imagine the wonders our country could know in America's 250th year.
Think of the marvels we could achieve if we simply set free the dreams of our people. Cures to the illnesses that have always plagued us are not too much to hope. American footprints on distant worlds are not too big a dream. Millions lifted from welfare to work is not too much to expect. And streets where mothers are safe from fear schools where children learn in peace, and jobs where Americans prosper and grow are not too much to ask.
When we have all of this, we will have made America greater than ever before, for all Americans. This is our vision. This is our mission. But we can only get there together. We are one people, with one destiny.
We all bleed the same blood. We all salute the same great American flag. And we are all made by the same God.
When we fulfill this vision, when we celebrate our 250 years of glorious freedom, we will look back on tonight as when this new chapter of American greatness began. The time for small thinking is over. The time for trivial fights is behind us. We just need the courage to share the dreams that fill our hearts, the bravery to express the hopes that stir our souls, and the confidence to turn those hopes and those dreams into action.
From now on, America will be empowered by our aspirations, not burdened by our fears, inspired by the future, not bound by failures of the past, and guided by a vision, not blinded by our doubts.
I am asking all citizens to embrace this renewal of the American spirit. I am asking all members of Congress to join me in dreaming big and bold and daring things for our country. I am asking everyone watching tonight to seize this moment. Believe in yourselves. Believe in your future. And believe, once more, in America.
Thank you, God bless you, and God bless the United States.
display(dbutils.fs.ls("dbfs:/"))
| path | name | size |
|---|---|---|
| dbfs:/FileStore/ | FileStore/ | 0.0 |
| dbfs:/_checkpoint/ | _checkpoint/ | 0.0 |
| dbfs:/databricks/ | databricks/ | 0.0 |
| dbfs:/databricks-datasets/ | databricks-datasets/ | 0.0 |
| dbfs:/databricks-results/ | databricks-results/ | 0.0 |
| dbfs:/datasets/ | datasets/ | 0.0 |
| dbfs:/digsum-dataframe.csv/ | digsum-dataframe.csv/ | 0.0 |
| dbfs:/ml/ | ml/ | 0.0 |
| dbfs:/mnt/ | mnt/ | 0.0 |
| dbfs:/mytmpdir-forUserTimeLine/ | mytmpdir-forUserTimeLine/ | 0.0 |
| dbfs:/results/ | results/ | 0.0 |
| dbfs:/test/ | test/ | 0.0 |
| dbfs:/tmp/ | tmp/ | 0.0 |
| dbfs:/tmpdir/ | tmpdir/ | 0.0 |
| dbfs:/user/ | user/ | 0.0 |
display(dbutils.fs.ls("dbfs:/datasets"))
| path | name | size |
|---|---|---|
| dbfs:/datasets/MEP/ | MEP/ | 0.0 |
| dbfs:/datasets/beijing/ | beijing/ | 0.0 |
| dbfs:/datasets/graphhopper/ | graphhopper/ | 0.0 |
| dbfs:/datasets/magellan/ | magellan/ | 0.0 |
| dbfs:/datasets/maps/ | maps/ | 0.0 |
| dbfs:/datasets/osm/ | osm/ | 0.0 |
| dbfs:/datasets/sou/ | sou/ | 0.0 |
| dbfs:/datasets/twitterAccountsOfInterest.csv | twitterAccountsOfInterest.csv | 32981.0 |
dbutils.fs.rm("dbfs:/datasets/sou/", recurse=true) //need to be done only if you want to delete such an existing directory, if any!
res2: Boolean = true
dbutils.fs.mkdirs("dbfs:/datasets/sou") //need not be done again if it already exists as desired!
res3: Boolean = true
display(dbutils.fs.ls("dbfs:/datasets"))
| path | name | size |
|---|---|---|
| dbfs:/datasets/MEP/ | MEP/ | 0.0 |
| dbfs:/datasets/beijing/ | beijing/ | 0.0 |
| dbfs:/datasets/graphhopper/ | graphhopper/ | 0.0 |
| dbfs:/datasets/magellan/ | magellan/ | 0.0 |
| dbfs:/datasets/maps/ | maps/ | 0.0 |
| dbfs:/datasets/osm/ | osm/ | 0.0 |
| dbfs:/datasets/sou/ | sou/ | 0.0 |
| dbfs:/datasets/twitterAccountsOfInterest.csv | twitterAccountsOfInterest.csv | 32981.0 |
pwd && ls
/databricks/driver
conf
derby.log
eventlogs
ganglia
logs
sou
sou.tgz
dbutils.fs.help
For more info about a method, use dbutils.fs.help("methodName").
In notebooks, you can also use the %fs shorthand to access DBFS. The %fs shorthand maps straightforwardly onto dbutils calls. For example, "%fs head --maxBytes=10000 /file/path" translates into "dbutils.fs.head("/file/path", maxBytes = 10000)".
fsutils
cp(from: String, to: String, recurse: boolean = false): boolean -> Copies a file or directory, possibly across FileSystemshead(file: String, maxBytes: int = 65536): String -> Returns up to the first 'maxBytes' bytes of the given file as a String encoded in UTF-8
ls(dir: String): Seq -> Lists the contents of a directory
mkdirs(dir: String): boolean -> Creates the given directory if it does not exist, also creating any necessary parent directories
mv(from: String, to: String, recurse: boolean = false): boolean -> Moves a file or directory, possibly across FileSystems
put(file: String, contents: String, overwrite: boolean = false): boolean -> Writes the given String out to a file, encoded in UTF-8
rm(dir: String, recurse: boolean = false): boolean -> Removes a file or directory
mount
mount(source: String, mountPoint: String, encryptionType: String = "", owner: String = null, extraConfigs: Map = Map.empty[String, String]): boolean -> Mounts the given source directory into DBFS at the given mount pointmounts: Seq -> Displays information about what is mounted within DBFS
refreshMounts: boolean -> Forces all machines in this cluster to refresh their mount cache, ensuring they receive the most recent information
unmount(mountPoint: String): boolean -> Deletes a DBFS mount point
dbutils.fs.cp("file:/databricks/driver/sou", "dbfs:/datasets/sou/",recurse=true)
res6: Boolean = true
display(dbutils.fs.ls("dbfs:/datasets/sou"))
| path | name | size |
|---|---|---|
| dbfs:/datasets/sou/17900108.txt | 17900108.txt | 6725.0 |
| dbfs:/datasets/sou/17901208.txt | 17901208.txt | 8427.0 |
| dbfs:/datasets/sou/17911025.txt | 17911025.txt | 14175.0 |
| dbfs:/datasets/sou/17921106.txt | 17921106.txt | 12736.0 |
| dbfs:/datasets/sou/17931203.txt | 17931203.txt | 11668.0 |
| dbfs:/datasets/sou/17941119.txt | 17941119.txt | 17615.0 |
| dbfs:/datasets/sou/17951208.txt | 17951208.txt | 12296.0 |
| dbfs:/datasets/sou/17961207.txt | 17961207.txt | 17340.0 |
| dbfs:/datasets/sou/17971122.txt | 17971122.txt | 12473.0 |
| dbfs:/datasets/sou/17981208.txt | 17981208.txt | 13394.0 |
| dbfs:/datasets/sou/17991203.txt | 17991203.txt | 9236.0 |
| dbfs:/datasets/sou/18001111.txt | 18001111.txt | 8382.0 |
| dbfs:/datasets/sou/18011208.txt | 18011208.txt | 19342.0 |
| dbfs:/datasets/sou/18021215.txt | 18021215.txt | 13003.0 |
| dbfs:/datasets/sou/18031017.txt | 18031017.txt | 14022.0 |
| dbfs:/datasets/sou/18041108.txt | 18041108.txt | 12652.0 |
| dbfs:/datasets/sou/18051203.txt | 18051203.txt | 17190.0 |
| dbfs:/datasets/sou/18061202.txt | 18061202.txt | 17135.0 |
| dbfs:/datasets/sou/18071027.txt | 18071027.txt | 14334.0 |
| dbfs:/datasets/sou/18081108.txt | 18081108.txt | 16225.0 |
| dbfs:/datasets/sou/18091129.txt | 18091129.txt | 11050.0 |
| dbfs:/datasets/sou/18101205.txt | 18101205.txt | 15028.0 |
| dbfs:/datasets/sou/18111105.txt | 18111105.txt | 13941.0 |
| dbfs:/datasets/sou/18121104.txt | 18121104.txt | 19615.0 |
| dbfs:/datasets/sou/18131207.txt | 18131207.txt | 19532.0 |
| dbfs:/datasets/sou/18140920.txt | 18140920.txt | 12632.0 |
| dbfs:/datasets/sou/18151205.txt | 18151205.txt | 19398.0 |
| dbfs:/datasets/sou/18161203.txt | 18161203.txt | 20331.0 |
| dbfs:/datasets/sou/18171212.txt | 18171212.txt | 26236.0 |
| dbfs:/datasets/sou/18181116.txt | 18181116.txt | 26445.0 |
| dbfs:/datasets/sou/18191207.txt | 18191207.txt | 27880.0 |
| dbfs:/datasets/sou/18201114.txt | 18201114.txt | 20503.0 |
| dbfs:/datasets/sou/18211203.txt | 18211203.txt | 34364.0 |
| dbfs:/datasets/sou/18221203.txt | 18221203.txt | 28154.0 |
| dbfs:/datasets/sou/18231202.txt | 18231202.txt | 38329.0 |
| dbfs:/datasets/sou/18241207.txt | 18241207.txt | 49869.0 |
| dbfs:/datasets/sou/18251206.txt | 18251206.txt | 53992.0 |
| dbfs:/datasets/sou/18261205.txt | 18261205.txt | 46482.0 |
| dbfs:/datasets/sou/18271204.txt | 18271204.txt | 42481.0 |
| dbfs:/datasets/sou/18281202.txt | 18281202.txt | 44202.0 |
| dbfs:/datasets/sou/18291208.txt | 18291208.txt | 62923.0 |
| dbfs:/datasets/sou/18301206.txt | 18301206.txt | 90641.0 |
| dbfs:/datasets/sou/18311206.txt | 18311206.txt | 42902.0 |
| dbfs:/datasets/sou/18321204.txt | 18321204.txt | 46879.0 |
| dbfs:/datasets/sou/18331203.txt | 18331203.txt | 46991.0 |
| dbfs:/datasets/sou/18341201.txt | 18341201.txt | 80364.0 |
| dbfs:/datasets/sou/18351207.txt | 18351207.txt | 64395.0 |
| dbfs:/datasets/sou/18361205.txt | 18361205.txt | 73306.0 |
| dbfs:/datasets/sou/18371205.txt | 18371205.txt | 68927.0 |
| dbfs:/datasets/sou/18381203.txt | 18381203.txt | 69880.0 |
| dbfs:/datasets/sou/18391202.txt | 18391202.txt | 80147.0 |
| dbfs:/datasets/sou/18401205.txt | 18401205.txt | 55025.0 |
| dbfs:/datasets/sou/18411207.txt | 18411207.txt | 48792.0 |
| dbfs:/datasets/sou/18421206.txt | 18421206.txt | 49788.0 |
| dbfs:/datasets/sou/18431206.txt | 18431206.txt | 47670.0 |
| dbfs:/datasets/sou/18441203.txt | 18441203.txt | 55494.0 |
| dbfs:/datasets/sou/18451202.txt | 18451202.txt | 95894.0 |
| dbfs:/datasets/sou/18461208.txt | 18461208.txt | 107852.0 |
| dbfs:/datasets/sou/18471207.txt | 18471207.txt | 96912.0 |
| dbfs:/datasets/sou/18481205.txt | 18481205.txt | 127557.0 |
| dbfs:/datasets/sou/18491204.txt | 18491204.txt | 46003.0 |
| dbfs:/datasets/sou/18501202.txt | 18501202.txt | 49823.0 |
| dbfs:/datasets/sou/18511202.txt | 18511202.txt | 79335.0 |
| dbfs:/datasets/sou/18521206.txt | 18521206.txt | 59438.0 |
| dbfs:/datasets/sou/18531205.txt | 18531205.txt | 58031.0 |
| dbfs:/datasets/sou/18541204.txt | 18541204.txt | 61917.0 |
| dbfs:/datasets/sou/18551231.txt | 18551231.txt | 70459.0 |
| dbfs:/datasets/sou/18561202.txt | 18561202.txt | 63906.0 |
| dbfs:/datasets/sou/18571208.txt | 18571208.txt | 82051.0 |
| dbfs:/datasets/sou/18581206.txt | 18581206.txt | 98523.0 |
| dbfs:/datasets/sou/18591219.txt | 18591219.txt | 74089.0 |
| dbfs:/datasets/sou/18601203.txt | 18601203.txt | 84283.0 |
| dbfs:/datasets/sou/18611203.txt | 18611203.txt | 41587.0 |
| dbfs:/datasets/sou/18621201.txt | 18621201.txt | 50008.0 |
| dbfs:/datasets/sou/18631208.txt | 18631208.txt | 37109.0 |
| dbfs:/datasets/sou/18641206.txt | 18641206.txt | 36201.0 |
| dbfs:/datasets/sou/18651204.txt | 18651204.txt | 54781.0 |
| dbfs:/datasets/sou/18661203.txt | 18661203.txt | 44152.0 |
| dbfs:/datasets/sou/18671203.txt | 18671203.txt | 71650.0 |
| dbfs:/datasets/sou/18681209.txt | 18681209.txt | 60650.0 |
| dbfs:/datasets/sou/18691206.txt | 18691206.txt | 46099.0 |
| dbfs:/datasets/sou/18701205.txt | 18701205.txt | 52113.0 |
| dbfs:/datasets/sou/18711204.txt | 18711204.txt | 38805.0 |
| dbfs:/datasets/sou/18721202.txt | 18721202.txt | 23984.0 |
| dbfs:/datasets/sou/18731201.txt | 18731201.txt | 60406.0 |
| dbfs:/datasets/sou/18741207.txt | 18741207.txt | 55136.0 |
| dbfs:/datasets/sou/18751207.txt | 18751207.txt | 73272.0 |
| dbfs:/datasets/sou/18761205.txt | 18761205.txt | 40873.0 |
| dbfs:/datasets/sou/18771203.txt | 18771203.txt | 48620.0 |
| dbfs:/datasets/sou/18781202.txt | 18781202.txt | 48552.0 |
| dbfs:/datasets/sou/18791201.txt | 18791201.txt | 71149.0 |
| dbfs:/datasets/sou/18801206.txt | 18801206.txt | 41294.0 |
| dbfs:/datasets/sou/18811206.txt | 18811206.txt | 24189.0 |
| dbfs:/datasets/sou/18821204.txt | 18821204.txt | 19065.0 |
| dbfs:/datasets/sou/18831204.txt | 18831204.txt | 23860.0 |
| dbfs:/datasets/sou/18841201.txt | 18841201.txt | 55230.0 |
| dbfs:/datasets/sou/18851208.txt | 18851208.txt | 121030.0 |
| dbfs:/datasets/sou/18861206.txt | 18861206.txt | 92873.0 |
| dbfs:/datasets/sou/18871206.txt | 18871206.txt | 31685.0 |
| dbfs:/datasets/sou/18881203.txt | 18881203.txt | 55460.0 |
| dbfs:/datasets/sou/18891203.txt | 18891203.txt | 77944.0 |
| dbfs:/datasets/sou/18901201.txt | 18901201.txt | 69588.0 |
| dbfs:/datasets/sou/18911209.txt | 18911209.txt | 96894.0 |
| dbfs:/datasets/sou/18921206.txt | 18921206.txt | 81825.0 |
| dbfs:/datasets/sou/18931203.txt | 18931203.txt | 76786.0 |
| dbfs:/datasets/sou/18941202.txt | 18941202.txt | 97793.0 |
| dbfs:/datasets/sou/18951207.txt | 18951207.txt | 89791.0 |
| dbfs:/datasets/sou/18961204.txt | 18961204.txt | 94943.0 |
| dbfs:/datasets/sou/18971206.txt | 18971206.txt | 72748.0 |
| dbfs:/datasets/sou/18981205.txt | 18981205.txt | 123819.0 |
| dbfs:/datasets/sou/18991205.txt | 18991205.txt | 93175.0 |
| dbfs:/datasets/sou/19001203.txt | 19001203.txt | 118487.0 |
| dbfs:/datasets/sou/19011203.txt | 19011203.txt | 115838.0 |
| dbfs:/datasets/sou/19021202.txt | 19021202.txt | 57671.0 |
| dbfs:/datasets/sou/19031207.txt | 19031207.txt | 90262.0 |
| dbfs:/datasets/sou/19041206.txt | 19041206.txt | 104031.0 |
| dbfs:/datasets/sou/19051205.txt | 19051205.txt | 147449.0 |
| dbfs:/datasets/sou/19061203.txt | 19061203.txt | 138165.0 |
| dbfs:/datasets/sou/19071203.txt | 19071203.txt | 161983.0 |
| dbfs:/datasets/sou/19081208.txt | 19081208.txt | 115609.0 |
| dbfs:/datasets/sou/19091207.txt | 19091207.txt | 84749.0 |
| dbfs:/datasets/sou/19101206.txt | 19101206.txt | 42598.0 |
| dbfs:/datasets/sou/19111205.txt | 19111205.txt | 143491.0 |
| dbfs:/datasets/sou/19121203.txt | 19121203.txt | 153124.0 |
| dbfs:/datasets/sou/19131202.txt | 19131202.txt | 20536.0 |
| dbfs:/datasets/sou/19141208.txt | 19141208.txt | 25441.0 |
| dbfs:/datasets/sou/19151207.txt | 19151207.txt | 44773.0 |
| dbfs:/datasets/sou/19161205.txt | 19161205.txt | 12773.0 |
| dbfs:/datasets/sou/19171204.txt | 19171204.txt | 22077.0 |
| dbfs:/datasets/sou/19181202.txt | 19181202.txt | 31400.0 |
| dbfs:/datasets/sou/19191202.txt | 19191202.txt | 28511.0 |
| dbfs:/datasets/sou/19201207.txt | 19201207.txt | 16119.0 |
| dbfs:/datasets/sou/19211206.txt | 19211206.txt | 34334.0 |
| dbfs:/datasets/sou/19221208.txt | 19221208.txt | 35419.0 |
| dbfs:/datasets/sou/19231206.txt | 19231206.txt | 41144.0 |
| dbfs:/datasets/sou/19241203.txt | 19241203.txt | 42503.0 |
| dbfs:/datasets/sou/19251208.txt | 19251208.txt | 66289.0 |
| dbfs:/datasets/sou/19261207.txt | 19261207.txt | 62608.0 |
| dbfs:/datasets/sou/19271206.txt | 19271206.txt | 54125.0 |
| dbfs:/datasets/sou/19281204.txt | 19281204.txt | 50110.0 |
| dbfs:/datasets/sou/19291203.txt | 19291203.txt | 68959.0 |
| dbfs:/datasets/sou/19301202.txt | 19301202.txt | 29041.0 |
| dbfs:/datasets/sou/19311208.txt | 19311208.txt | 36649.0 |
| dbfs:/datasets/sou/19321206.txt | 19321206.txt | 26421.0 |
| dbfs:/datasets/sou/19340103.txt | 19340103.txt | 13545.0 |
| dbfs:/datasets/sou/19350104.txt | 19350104.txt | 21221.0 |
| dbfs:/datasets/sou/19360103.txt | 19360103.txt | 22300.0 |
| dbfs:/datasets/sou/19370106.txt | 19370106.txt | 16738.0 |
| dbfs:/datasets/sou/19380103.txt | 19380103.txt | 28069.0 |
| dbfs:/datasets/sou/19390104.txt | 19390104.txt | 22563.0 |
| dbfs:/datasets/sou/19400103.txt | 19400103.txt | 18722.0 |
| dbfs:/datasets/sou/19410106.txt | 19410106.txt | 19386.0 |
| dbfs:/datasets/sou/19420106.txt | 19420106.txt | 19911.0 |
| dbfs:/datasets/sou/19430107.txt | 19430107.txt | 26314.0 |
| dbfs:/datasets/sou/19440111.txt | 19440111.txt | 22151.0 |
| dbfs:/datasets/sou/19450106.txt | 19450106.txt | 48891.0 |
| dbfs:/datasets/sou/19460121.txt | 19460121.txt | 174651.0 |
| dbfs:/datasets/sou/19470106.txt | 19470106.txt | 37406.0 |
| dbfs:/datasets/sou/19480107.txt | 19480107.txt | 30550.0 |
| dbfs:/datasets/sou/19490105.txt | 19490105.txt | 20792.0 |
| dbfs:/datasets/sou/19500104.txt | 19500104.txt | 30423.0 |
| dbfs:/datasets/sou/19510108.txt | 19510108.txt | 22924.0 |
| dbfs:/datasets/sou/19520109.txt | 19520109.txt | 30228.0 |
| dbfs:/datasets/sou/19530107.txt | 19530107.txt | 56767.0 |
| dbfs:/datasets/sou/19530202.txt | 19530202.txt | 43620.0 |
| dbfs:/datasets/sou/19540107.txt | 19540107.txt | 37843.0 |
| dbfs:/datasets/sou/19550106.txt | 19550106.txt | 46532.0 |
| dbfs:/datasets/sou/19560105.txt | 19560105.txt | 52138.0 |
| dbfs:/datasets/sou/19570110.txt | 19570110.txt | 25846.0 |
| dbfs:/datasets/sou/19580109.txt | 19580109.txt | 30344.0 |
| dbfs:/datasets/sou/19590109.txt | 19590109.txt | 30145.0 |
| dbfs:/datasets/sou/19600107.txt | 19600107.txt | 35099.0 |
| dbfs:/datasets/sou/19610112.txt | 19610112.txt | 40396.0 |
| dbfs:/datasets/sou/19610130.txt | 19610130.txt | 31641.0 |
| dbfs:/datasets/sou/19620111.txt | 19620111.txt | 39488.0 |
| dbfs:/datasets/sou/19630114.txt | 19630114.txt | 31666.0 |
| dbfs:/datasets/sou/19640108.txt | 19640108.txt | 18659.0 |
| dbfs:/datasets/sou/19650104.txt | 19650104.txt | 25389.0 |
| dbfs:/datasets/sou/19660112.txt | 19660112.txt | 30570.0 |
| dbfs:/datasets/sou/19670110.txt | 19670110.txt | 41668.0 |
| dbfs:/datasets/sou/19680117.txt | 19680117.txt | 28834.0 |
| dbfs:/datasets/sou/19690114.txt | 19690114.txt | 23634.0 |
| dbfs:/datasets/sou/19700122.txt | 19700122.txt | 25408.0 |
| dbfs:/datasets/sou/19710122.txt | 19710122.txt | 25793.0 |
| dbfs:/datasets/sou/19720120.txt | 19720120.txt | 23099.0 |
| dbfs:/datasets/sou/19730202.txt | 19730202.txt | 9844.0 |
| dbfs:/datasets/sou/19740130.txt | 19740130.txt | 29231.0 |
| dbfs:/datasets/sou/19750115.txt | 19750115.txt | 24801.0 |
| dbfs:/datasets/sou/19760119.txt | 19760119.txt | 29731.0 |
| dbfs:/datasets/sou/19770112.txt | 19770112.txt | 27923.0 |
| dbfs:/datasets/sou/19780119.txt | 19780119.txt | 26564.0 |
| dbfs:/datasets/sou/19790125.txt | 19790125.txt | 19544.0 |
| dbfs:/datasets/sou/19800121.txt | 19800121.txt | 20124.0 |
| dbfs:/datasets/sou/19810116.txt | 19810116.txt | 217980.0 |
| dbfs:/datasets/sou/19820126.txt | 19820126.txt | 31166.0 |
| dbfs:/datasets/sou/19830125.txt | 19830125.txt | 33255.0 |
| dbfs:/datasets/sou/19840125.txt | 19840125.txt | 29705.0 |
| dbfs:/datasets/sou/19850206.txt | 19850206.txt | 25364.0 |
| dbfs:/datasets/sou/19860204.txt | 19860204.txt | 20449.0 |
| dbfs:/datasets/sou/19870127.txt | 19870127.txt | 22334.0 |
| dbfs:/datasets/sou/19880125.txt | 19880125.txt | 28565.0 |
| dbfs:/datasets/sou/19890209.txt | 19890209.txt | 27855.0 |
| dbfs:/datasets/sou/19900131.txt | 19900131.txt | 21434.0 |
| dbfs:/datasets/sou/19910129.txt | 19910129.txt | 22433.0 |
| dbfs:/datasets/sou/19920128.txt | 19920128.txt | 26644.0 |
| dbfs:/datasets/sou/19930217.txt | 19930217.txt | 39255.0 |
| dbfs:/datasets/sou/19940125.txt | 19940125.txt | 42320.0 |
| dbfs:/datasets/sou/19950124.txt | 19950124.txt | 51325.0 |
| dbfs:/datasets/sou/19960123.txt | 19960123.txt | 36386.0 |
| dbfs:/datasets/sou/19970204.txt | 19970204.txt | 39038.0 |
| dbfs:/datasets/sou/19980127.txt | 19980127.txt | 42255.0 |
| dbfs:/datasets/sou/19990119.txt | 19990119.txt | 43592.0 |
| dbfs:/datasets/sou/20000127.txt | 20000127.txt | 44244.0 |
| dbfs:/datasets/sou/20010227.txt | 20010227.txt | 25330.0 |
| dbfs:/datasets/sou/20010920.txt | 20010920.txt | 17383.0 |
| dbfs:/datasets/sou/20020129.txt | 20020129.txt | 22653.0 |
| dbfs:/datasets/sou/20030128.txt | 20030128.txt | 31878.0 |
| dbfs:/datasets/sou/20040120.txt | 20040120.txt | 30611.0 |
| dbfs:/datasets/sou/20050202.txt | 20050202.txt | 29875.0 |
| dbfs:/datasets/sou/20060131.txt | 20060131.txt | 31449.0 |
| dbfs:/datasets/sou/20070123.txt | 20070123.txt | 31998.0 |
| dbfs:/datasets/sou/20080128.txt | 20080128.txt | 33830.0 |
| dbfs:/datasets/sou/20090224.txt | 20090224.txt | 33640.0 |
| dbfs:/datasets/sou/20100127.txt | 20100127.txt | 40980.0 |
| dbfs:/datasets/sou/20110125.txt | 20110125.txt | 39582.0 |
| dbfs:/datasets/sou/20120124.txt | 20120124.txt | 40338.0 |
| dbfs:/datasets/sou/20130212.txt | 20130212.txt | 37815.0 |
| dbfs:/datasets/sou/20140128.txt | 20140128.txt | 39625.0 |
| dbfs:/datasets/sou/20150120.txt | 20150120.txt | 38528.0 |
| dbfs:/datasets/sou/20160112.txt | 20160112.txt | 31083.0 |
| dbfs:/datasets/sou/20170228.txt | 20170228.txt | 29323.0 |
display(dbutils.fs.ls("dbfs:/datasets/"))
| path | name | size |
|---|---|---|
| dbfs:/datasets/MEP/ | MEP/ | 0.0 |
| dbfs:/datasets/beijing/ | beijing/ | 0.0 |
| dbfs:/datasets/graphhopper/ | graphhopper/ | 0.0 |
| dbfs:/datasets/magellan/ | magellan/ | 0.0 |
| dbfs:/datasets/maps/ | maps/ | 0.0 |
| dbfs:/datasets/osm/ | osm/ | 0.0 |
| dbfs:/datasets/sou/ | sou/ | 0.0 |
| dbfs:/datasets/twitterAccountsOfInterest.csv | twitterAccountsOfInterest.csv | 32981.0 |
val sou17900108 = sc.textFile("dbfs:/datasets/sou/17900108.txt")
sou17900108: org.apache.spark.rdd.RDD[String] = dbfs:/datasets/sou/17900108.txt MapPartitionsRDD[51] at textFile at command-3718738704207872:1
sou17900108.take(5)
res11: Array[String] = Array("George Washington ", "", "January 8, 1790 ", "Fellow-Citizens of the Senate and House of Representatives: ", "I embrace with great satisfaction the opportunity which now presents itself of congratulating you on the present favorable prospects of our public affairs. The recent accession of the important state of North Carolina to the Constitution of the United States (of which official information has been received), the rising credit and respectability of our country, the general and increasing good will toward the government of the Union, and the concord, peace, and plenty with which we are blessed are circumstances auspicious in an eminent degree to our national prosperity. ")
sou17900108.collect
res12: Array[String] = Array("George Washington ", "", "January 8, 1790 ", "Fellow-Citizens of the Senate and House of Representatives: ", "I embrace with great satisfaction the opportunity which now presents itself of congratulating you on the present favorable prospects of our public affairs. The recent accession of the important state of North Carolina to the Constitution of the United States (of which official information has been received), the rising credit and respectability of our country, the general and increasing good will toward the government of the Union, and the concord, peace, and plenty with which we are blessed are circumstances auspicious in an eminent degree to our national prosperity. ", "In resuming your consultations for the general good you can not but derive encouragement from the reflection that the measures of the last session have been as satisfactory to your constituents as the novelty and difficulty of the work allowed you to hope. Still further to realize their expectations and to secure the blessings which a gracious Providence has placed within our reach will in the course of the present important session call for the cool and deliberate exertion of your patriotism, firmness, and wisdom. ", "Among the many interesting objects which will engage your attention that of providing for the common defense will merit particular regard. To be prepared for war is one of the most effectual means of preserving peace. ", "A free people ought not only to be armed, but disciplined; to which end a uniform and well-digested plan is requisite; and their safety and interest require that they should promote such manufactories as tend to render them independent of others for essential, particularly military, supplies. ", "The proper establishment of the troops which may be deemed indispensable will be entitled to mature consideration. In the arrangements which may be made respecting it it will be of importance to conciliate the comfortable support of the officers and soldiers with a due regard to economy. ", "There was reason to hope that the pacific measures adopted with regard to certain hostile tribes of Indians would have relieved the inhabitants of our southern and western frontiers from their depredations, but you will perceive from the information contained in the papers which I shall direct to be laid before you (comprehending a communication from the Commonwealth of Virginia) that we ought to be prepared to afford protection to those parts of the Union, and, if necessary, to punish aggressors. ", "The interests of the United States require that our intercourse with other nations should be facilitated by such provisions as will enable me to fulfill my duty in that respect in the manner which circumstances may render most conducive to the public good, and to this end that the compensation to be made to the persons who may be employed should, according to the nature of their appointments, be defined by law, and a competent fund designated for defraying the expenses incident to the conduct of foreign affairs. ", "Various considerations also render it expedient that the terms on which foreigners may be admitted to the rights of citizens should be speedily ascertained by a uniform rule of naturalization. ", "Uniformity in the currency, weights, and measures of the United States is an object of great importance, and will, I am persuaded, be duly attended to. ", "The advancement of agriculture, commerce, and manufactures by all proper means will not, I trust, need recommendation; but I can not forbear intimating to you the expediency of giving effectual encouragement as well to the introduction of new and useful inventions from abroad as to the exertions of skill and genius in producing them at home, and of facilitating the intercourse between the distant parts of our country by a due attention to the post-office and post-roads. ", "Nor am I less persuaded that you will agree with me in opinion that there is nothing which can better deserve your patronage than the promotion of science and literature. Knowledge is in every country the surest basis of public happiness. In one in which the measures of government receive their impressions so immediately from the sense of the community as in ours it is proportionably essential. ", "To the security of a free constitution it contributes in various ways--by convincing those who are intrusted with the public administration that every valuable end of government is best answered by the enlightened confidence of the people, and by teaching the people themselves to know and to value their own rights; to discern and provide against invasions of them; to distinguish between oppression and the necessary exercise of lawful authority; between burthens proceeding from a disregard to their convenience and those resulting from the inevitable exigencies of society; to discriminate the spirit of liberty from that of licentiousness-- cherishing the first, avoiding the last--and uniting a speedy but temperate vigilance against encroachments, with an inviolable respect to the laws. ", "Whether this desirable object will be best promoted by affording aids to seminaries of learning already established, by the institution of a national university, or by any other expedients will be well worthy of a place in the deliberations of the legislature. ", "Gentlemen of the House of Representatives: ", "I saw with peculiar pleasure at the close of the last session the resolution entered into by you expressive of your opinion that an adequate provision for the support of the public credit is a matter of high importance to the national honor and prosperity. In this sentiment I entirely concur; and to a perfect confidence in your best endeavors to devise such a provision as will be truly with the end I add an equal reliance on the cheerful cooperation of the other branch of the legislature. ", "It would be superfluous to specify inducements to a measure in which the character and interests of the United States are so obviously so deeply concerned, and which has received so explicit a sanction from your declaration. ", "Gentlemen of the Senate and House of Representatives: ", "I have directed the proper officers to lay before you, respectively, such papers and estimates as regard the affairs particularly recommended to your consideration, and necessary to convey to you that information of the state of the Union which it is my duty to afford. ", The welfare of our country is the great object to which our cares and efforts ought to be directed, and I shall derive great satisfaction from a cooperation with you in the pleasing though arduous task of insuring to our fellow citizens the blessings which they have a right to expect from a free, efficient, and equal government.)
sou17900108.takeOrdered(5)
res13: Array[String] = Array("", "A free people ought not only to be armed, but disciplined; to which end a uniform and well-digested plan is requisite; and their safety and interest require that they should promote such manufactories as tend to render them independent of others for essential, particularly military, supplies. ", "Among the many interesting objects which will engage your attention that of providing for the common defense will merit particular regard. To be prepared for war is one of the most effectual means of preserving peace. ", "Fellow-Citizens of the Senate and House of Representatives: ", "Gentlemen of the House of Representatives: ")
val souAll = sc.wholeTextFiles("dbfs:/datasets/sou/*.txt")
souAll: org.apache.spark.rdd.RDD[(String, String)] = dbfs:/datasets/sou/*.txt MapPartitionsRDD[53] at wholeTextFiles at command-3718738704207876:1
souAll.count // should be 231
res14: Long = 231
souAll.take(2)
res15: Array[(String, String)] =
Array((dbfs:/datasets/sou/17900108.txt,"George Washington
January 8, 1790
Fellow-Citizens of the Senate and House of Representatives:
I embrace with great satisfaction the opportunity which now presents itself of congratulating you on the present favorable prospects of our public affairs. The recent accession of the important state of North Carolina to the Constitution of the United States (of which official information has been received), the rising credit and respectability of our country, the general and increasing good will toward the government of the Union, and the concord, peace, and plenty with which we are blessed are circumstances auspicious in an eminent degree to our national prosperity.
In resuming your consultations for the general good you can not but derive encouragement from the reflection that the measures of the last session have been as satisfactory to your constituents as the novelty and difficulty of the work allowed you to hope. Still further to realize their expectations and to secure the blessings which a gracious Providence has placed within our reach will in the course of the present important session call for the cool and deliberate exertion of your patriotism, firmness, and wisdom.
Among the many interesting objects which will engage your attention that of providing for the common defense will merit particular regard. To be prepared for war is one of the most effectual means of preserving peace.
A free people ought not only to be armed, but disciplined; to which end a uniform and well-digested plan is requisite; and their safety and interest require that they should promote such manufactories as tend to render them independent of others for essential, particularly military, supplies.
The proper establishment of the troops which may be deemed indispensable will be entitled to mature consideration. In the arrangements which may be made respecting it it will be of importance to conciliate the comfortable support of the officers and soldiers with a due regard to economy.
There was reason to hope that the pacific measures adopted with regard to certain hostile tribes of Indians would have relieved the inhabitants of our southern and western frontiers from their depredations, but you will perceive from the information contained in the papers which I shall direct to be laid before you (comprehending a communication from the Commonwealth of Virginia) that we ought to be prepared to afford protection to those parts of the Union, and, if necessary, to punish aggressors.
The interests of the United States require that our intercourse with other nations should be facilitated by such provisions as will enable me to fulfill my duty in that respect in the manner which circumstances may render most conducive to the public good, and to this end that the compensation to be made to the persons who may be employed should, according to the nature of their appointments, be defined by law, and a competent fund designated for defraying the expenses incident to the conduct of foreign affairs.
Various considerations also render it expedient that the terms on which foreigners may be admitted to the rights of citizens should be speedily ascertained by a uniform rule of naturalization.
Uniformity in the currency, weights, and measures of the United States is an object of great importance, and will, I am persuaded, be duly attended to.
The advancement of agriculture, commerce, and manufactures by all proper means will not, I trust, need recommendation; but I can not forbear intimating to you the expediency of giving effectual encouragement as well to the introduction of new and useful inventions from abroad as to the exertions of skill and genius in producing them at home, and of facilitating the intercourse between the distant parts of our country by a due attention to the post-office and post-roads.
Nor am I less persuaded that you will agree with me in opinion that there is nothing which can better deserve your patronage than the promotion of science and literature. Knowledge is in every country the surest basis of public happiness. In one in which the measures of government receive their impressions so immediately from the sense of the community as in ours it is proportionably essential.
To the security of a free constitution it contributes in various ways--by convincing those who are intrusted with the public administration that every valuable end of government is best answered by the enlightened confidence of the people, and by teaching the people themselves to know and to value their own rights; to discern and provide against invasions of them; to distinguish between oppression and the necessary exercise of lawful authority; between burthens proceeding from a disregard to their convenience and those resulting from the inevitable exigencies of society; to discriminate the spirit of liberty from that of licentiousness-- cherishing the first, avoiding the last--and uniting a speedy but temperate vigilance against encroachments, with an inviolable respect to the laws.
Whether this desirable object will be best promoted by affording aids to seminaries of learning already established, by the institution of a national university, or by any other expedients will be well worthy of a place in the deliberations of the legislature.
Gentlemen of the House of Representatives:
I saw with peculiar pleasure at the close of the last session the resolution entered into by you expressive of your opinion that an adequate provision for the support of the public credit is a matter of high importance to the national honor and prosperity. In this sentiment I entirely concur; and to a perfect confidence in your best endeavors to devise such a provision as will be truly with the end I add an equal reliance on the cheerful cooperation of the other branch of the legislature.
It would be superfluous to specify inducements to a measure in which the character and interests of the United States are so obviously so deeply concerned, and which has received so explicit a sanction from your declaration.
Gentlemen of the Senate and House of Representatives:
I have directed the proper officers to lay before you, respectively, such papers and estimates as regard the affairs particularly recommended to your consideration, and necessary to convey to you that information of the state of the Union which it is my duty to afford.
The welfare of our country is the great object to which our cares and efforts ought to be directed, and I shall derive great satisfaction from a cooperation with you in the pleasing though arduous task of insuring to our fellow citizens the blessings which they have a right to expect from a free, efficient, and equal government.
"), (dbfs:/datasets/sou/17901208.txt,"George Washington
December 8, 1790
Fellow-Citizens of the Senate and House of Representatives:
In meeting you again I feel much satisfaction in being able to repeat my congratulations on the favorable prospects which continue to distinguish our public affairs. The abundant fruits of another year have blessed our country with plenty and with the means of a flourishing commerce.
The progress of public credit is witnessed by a considerable rise of American stock abroad as well as at home, and the revenues allotted for this and other national purposes have been productive beyond the calculations by which they were regulated. This latter circumstance is the more pleasing, as it is not only a proof of the fertility of our resources, but as it assures us of a further increase of the national respectability and credit, and, let me add, as it bears an honorable testimony to the patriotism and integrity of the mercantile and marine part of our citizens. The punctuality of the former in discharging their engagements has been exemplary.
In conformity to the powers vested in me by acts of the last session, a loan of 3,000,000 florins, toward which some provisional measures had previously taken place, has been completed in Holland. As well the celerity with which it has been filled as the nature of the terms (considering the more than ordinary demand for borrowing created by the situation of Europe) give a reasonable hope that the further execution of those powers may proceed with advantage and success. The Secretary of the Treasury has my directions to communicate such further particulars as may be requisite for more precise information.
Since your last sessions I have received communications by which it appears that the district of Kentucky, at present a part of Virginia, has concurred in certain propositions contained in a law of that State, in consequence of which the district is to become a distinct member of the Union, in case the requisite sanction of Congress be added. For this sanction application is now made. I shall cause the papers on this very transaction to be laid before you.
The liberality and harmony with which it has been conducted will be found to do great honor to both the parties, and the sentiments of warm attachment to the Union and its present Government expressed by our fellow citizens of Kentucky can not fail to add an affectionate concern for their particular welfare to the great national impressions under which you will decide on the case submitted to you.
It has been heretofore known to Congress that frequent incursions have been made on our frontier settlements by certain banditti of Indians from the northwest side of the Ohio. These, with some of the tribes dwelling on and near the Wabash, have of late been particularly active in their depredations, and being emboldened by the impunity of their crimes and aided by such parts of the neighboring tribes as could be seduced to join in their hostilities or afford them a retreat for their prisoners and plunder, they have, instead of listening to the humane invitations and overtures made on the part of the United States, renewed their violences with fresh alacrity and greater effect. The lives of a number of valuable citizens have thus been sacrificed, and some of them under circumstances peculiarly shocking, whilst others have been carried into a deplorable captivity.
These aggravated provocations rendered it essential to the safety of the Western settlements that the aggressors should be made sensible that the Government of the Union is not less capable of punishing their crimes than it is disposed to respect their rights and reward their attachments. As this object could not be effected by defensive measures, it became necessary to put in force the act which empowers the President to call out the militia for the protection of the frontiers, and I have accordingly authorized an expedition in which the regular troops in that quarter are combined with such drafts of militia as were deemed sufficient. The event of the measure is yet unknown to me. The Secretary of War is directed to lay before you a statement of the information on which it is founded, as well as an estimate of the expense with which it will be attended.
The disturbed situation of Europe, and particularly the critical posture of the great maritime powers, whilst it ought to make us the more thankful for the general peace and security enjoyed by the United States, reminds us at the same time of the circumspection with which it becomes us to preserve these blessings. It requires also that we should not overlook the tendency of a war, and even of preparations for a war, among the nations most concerned in active commerce with this country to abridge the means, and thereby at least enhance the price, of transporting its valuable productions to their markets. I recommend it to your serious reflections how far and in what mode it may be expedient to guard against embarrassments from these contingencies by such encouragements to our own navigation as will render our commerce and agriculture less dependent on foreign bottoms, which may fail us in the very moments most interesting to both of these great objects. Our fisheries and the transportation of our own produce offer us abundant means for guarding ourselves against this evil.
Your attention seems to be not less due to that particular branch of our trade which belongs to the Mediterranean. So many circumstances unite in rendering the present state of it distressful to us that you will not think any deliberations misemployed which may lead to its relief and protection.
The laws you have already passed for the establishment of a judiciary system have opened the doors of justice to all descriptions of persons. You will consider in your wisdom whether improvements in that system may yet be made, and particularly whether an uniform process of execution on sentences issuing from the Federal courts be not desirable through all the States.
The patronage of our commerce, of our merchants and sea men, has called for the appointment of consuls in foreign countries. It seems expedient to regulate by law the exercise of that jurisdiction and those functions which are permitted them, either by express convention or by a friendly indulgence, in the places of their residence. The consular convention, too, with His Most Christian Majesty has stipulated in certain cases the aid of the national authority to his consuls established here. Some legislative provision is requisite to carry these stipulations into full effect.
The establishment of the militia, of a mint, of standards of weights and measures, of the post office and post roads are subjects which I presume you will resume of course, and which are abundantly urged by their own importance.
Gentlemen of the House of Representatives:
The sufficiency of the revenues you have established for the objects to which they are appropriated leaves no doubt that the residuary provisions will be commensurate to the other objects for which the public faith stands now pledged. Allow me, moreover, to hope that it will be a favorite policy with you, not merely to secure a payment of the interest of the debt funded, but as far and as fast as the growing resources of the country will permit to exonerate it of the principal itself. The appropriation you have made of the Western land explains your dispositions on this subject, and I am persuaded that the sooner that valuable fund can be made to contribute, along with the other means, to the actual reduction of the public debt the more salutary will the measure be to every public interest, as well as the more satisfactory to our constituents.
Gentlemen of the Senate and House of Representatives:
In pursuing the various and weighty business of the present session I indulge the fullest persuasion that your consultation will be equally marked with wisdom and animated by the love of your country. In whatever belongs to my duty you shall have all the cooperation which an undiminished zeal for its welfare can inspire. It will be happy for us both, and our best reward, if, by a successful administration of our respective trusts, we can make the established Government more and more instrumental in promoting the good of our fellow citizens, and more and more the object of their attachment and confidence.
GO. WASHINGTON
"))
// This is a sequence of functions that can be accessed by other notebooks using the following %run command:
// %run "/scalable-data-science/xtraResources/support/sdsFunctions"
//This allows easy embedding of publicly available information into any other notebook
//when viewing in git-book just ignore this block - you may have to manually chase the URL in frameIt("URL").
//Example usage:
// displayHTML(frameIt("https://en.wikipedia.org/wiki/Latent_Dirichlet_allocation#Topics_in_LDA",250))
def frameIt( u:String, h:Int ) : String = {
"""<iframe
src=""""+ u+""""
width="95%" height="""" + h + """"
sandbox>
<p>
<a href="http://spark.apache.org/docs/latest/index.html">
Fallback link for browsers that, unlikely, don't support frames
</a>
</p>
</iframe>"""
}
frameIt: (u: String, h: Int)String
A visual guide to the transformations and actions on RDDs
Transformations
- filter
- flatMap
- groupBy
- groupByKey
- reduceByKey
- mapPartitions
- mapPartitionsWithIndex
- sample
- union
- join
- distinct
- coalesce
- keyBy
- partitionBy
Actions
Volunteers please... make PRs with the programing guide completed for ALL functions for which PNG images exist in: * https://github.com/lamastex/scalable-data-science/tree/master/db/visualapi/med

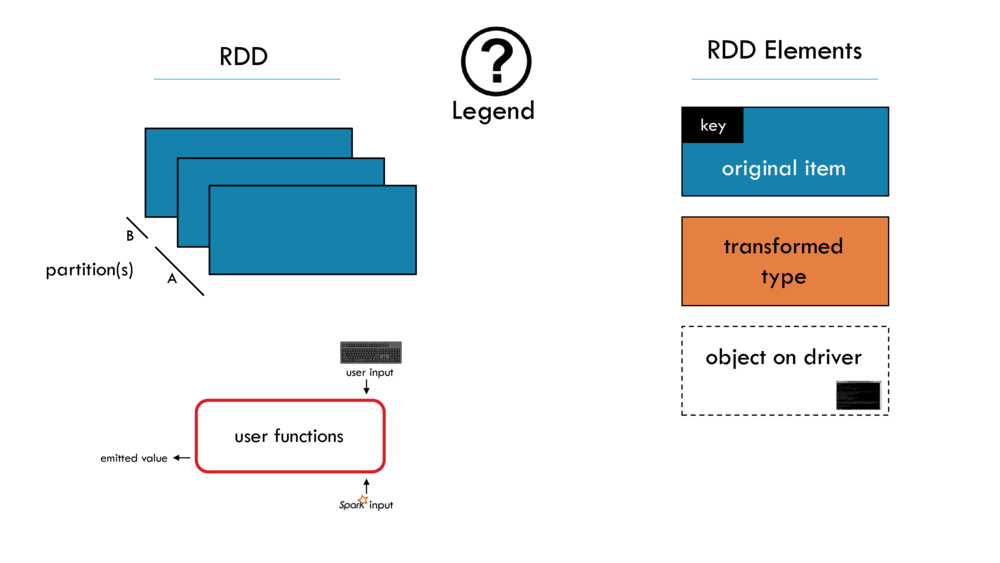
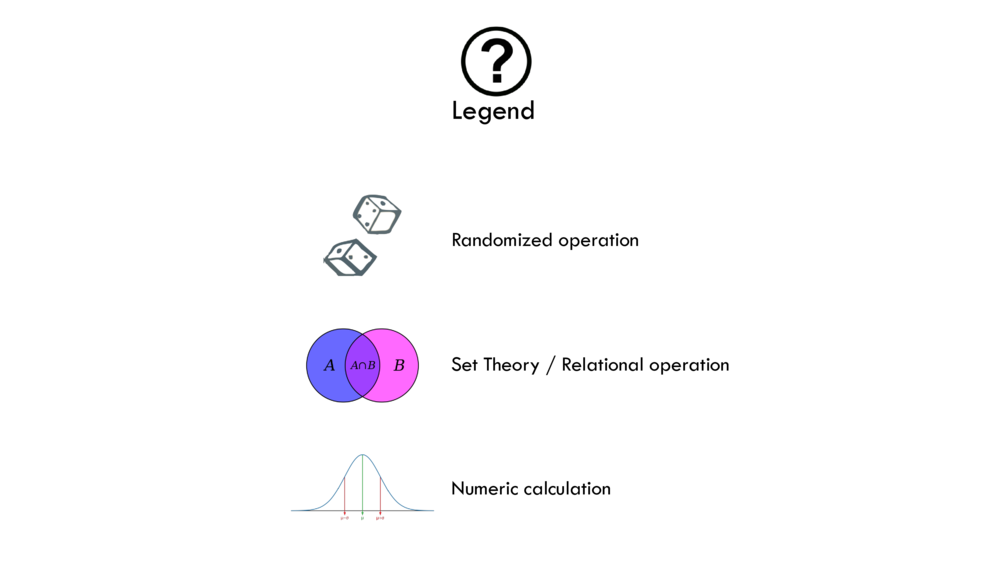
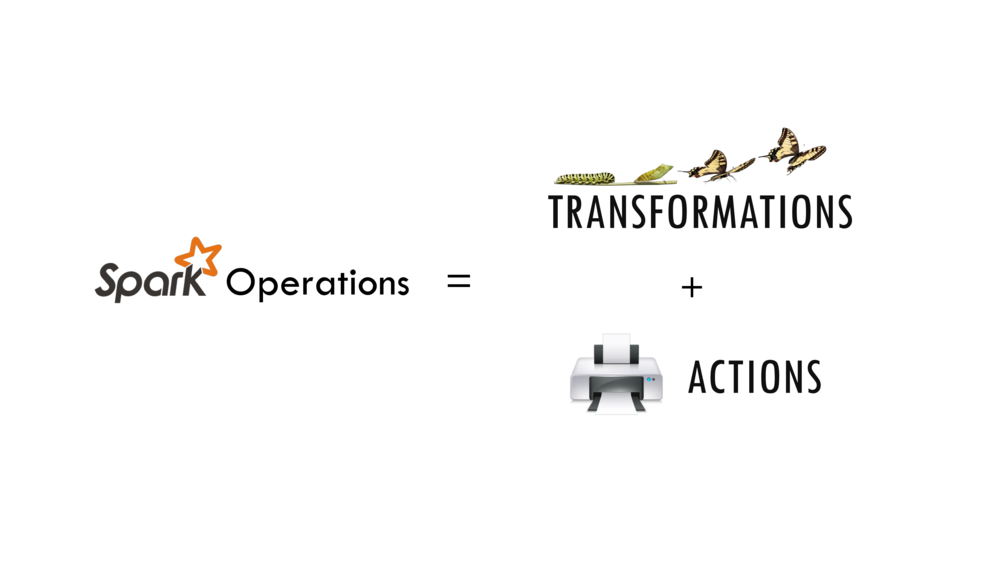
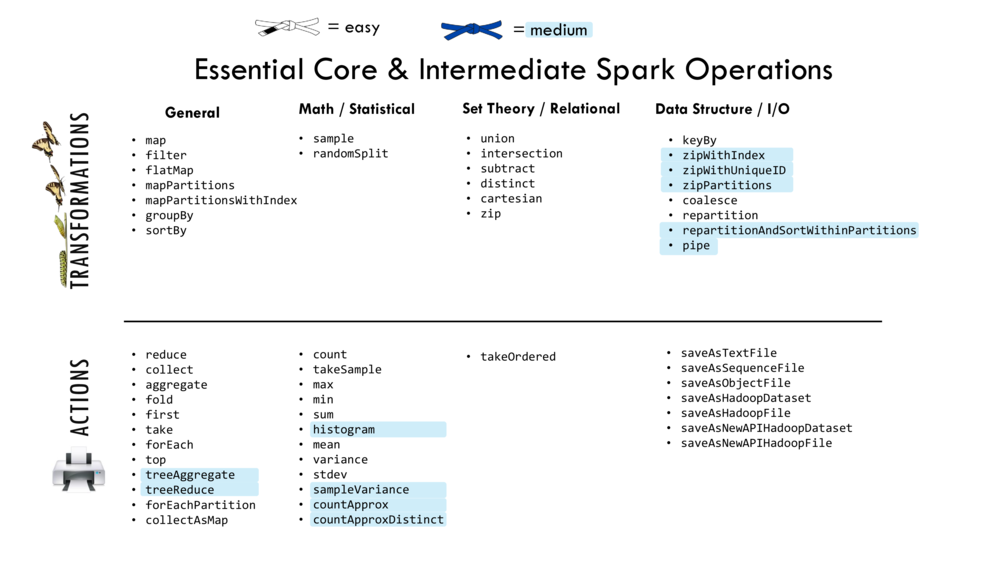
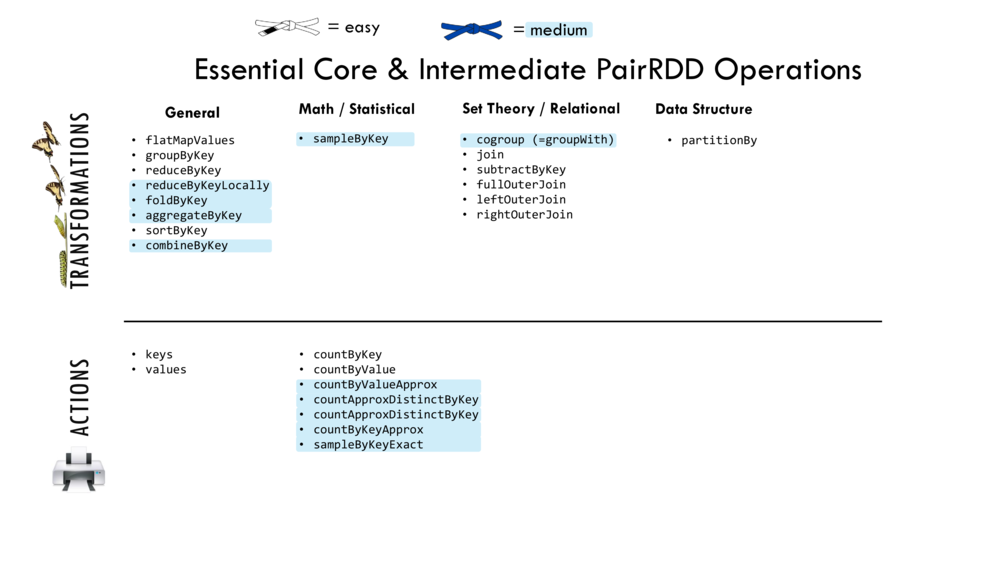

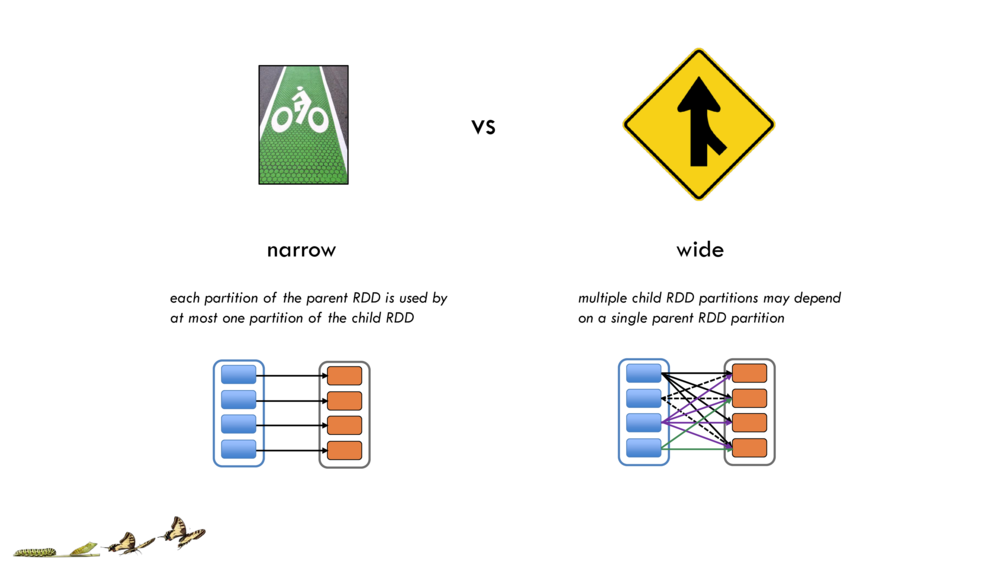
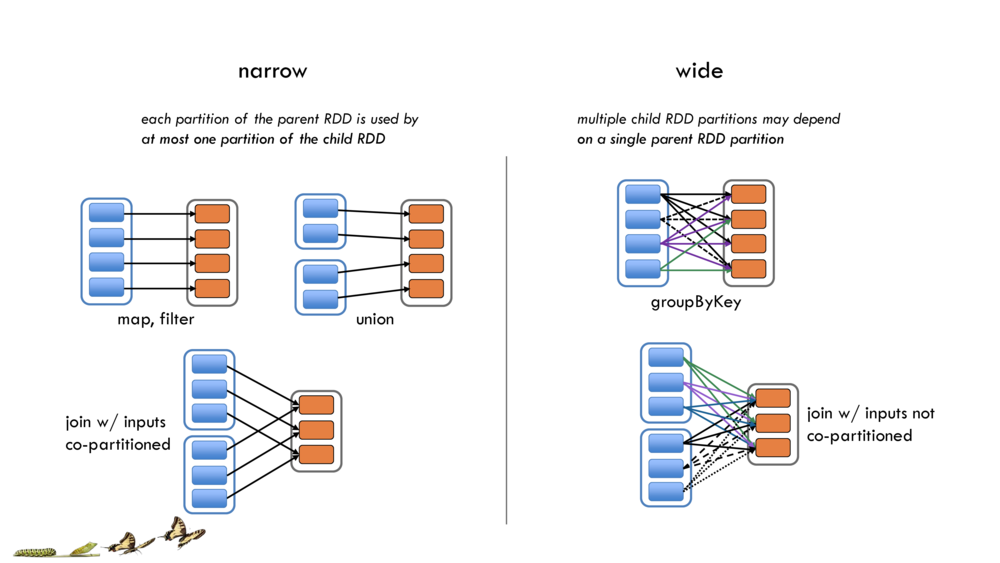

Continue from:

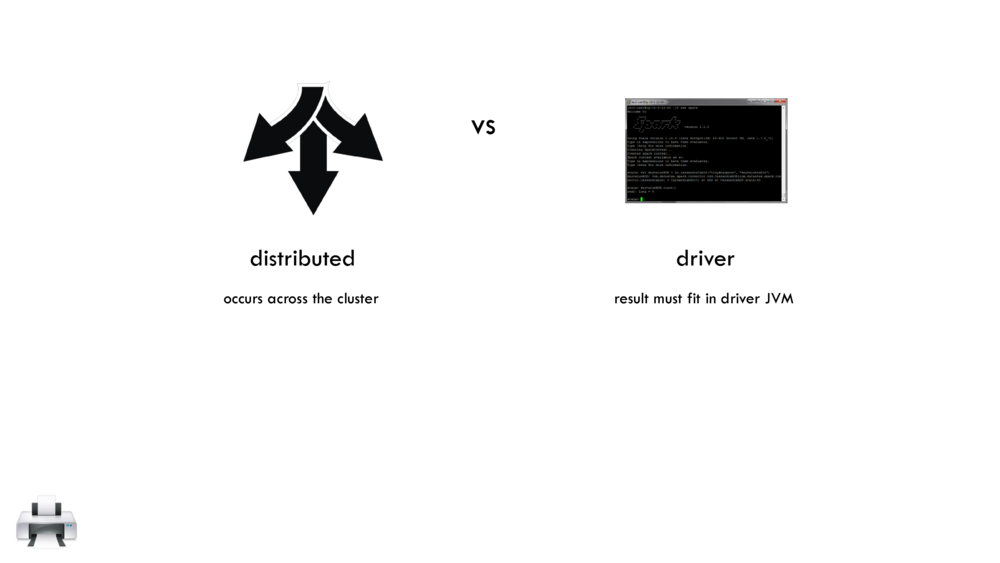
Continue from:
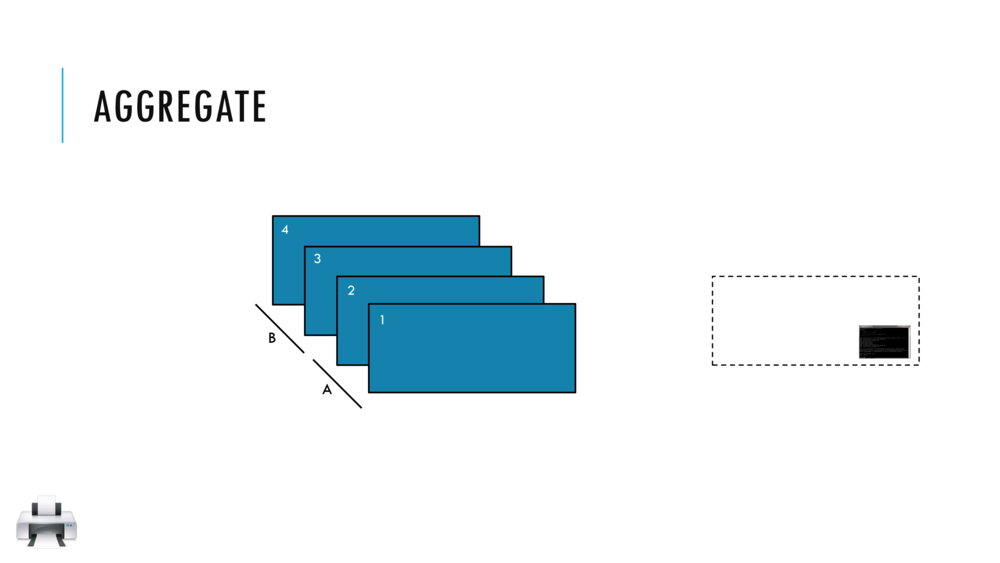
collect
Return all items in the RDD to the driver in a single list.
Let us look at the legend and overview of the visual RDD Api.
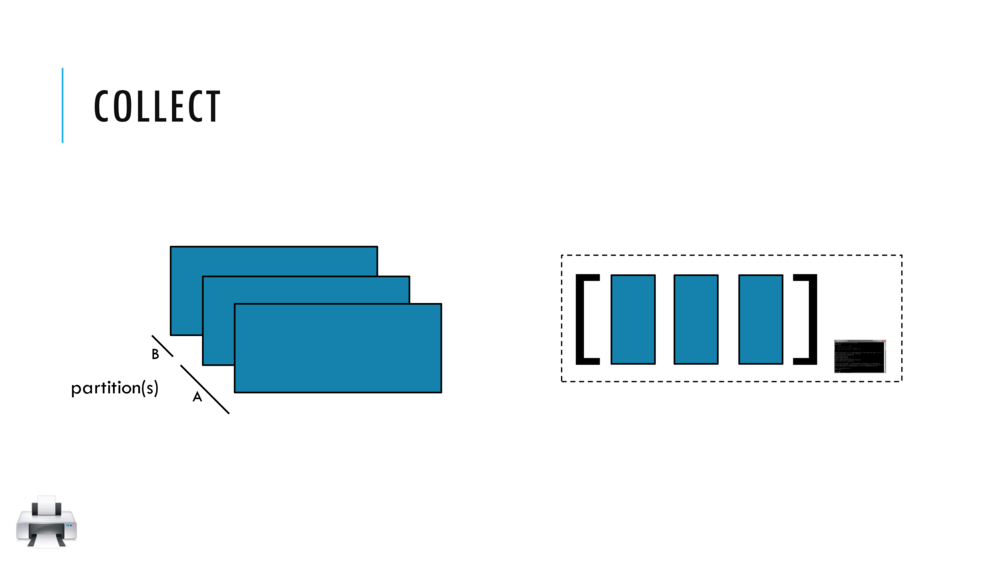
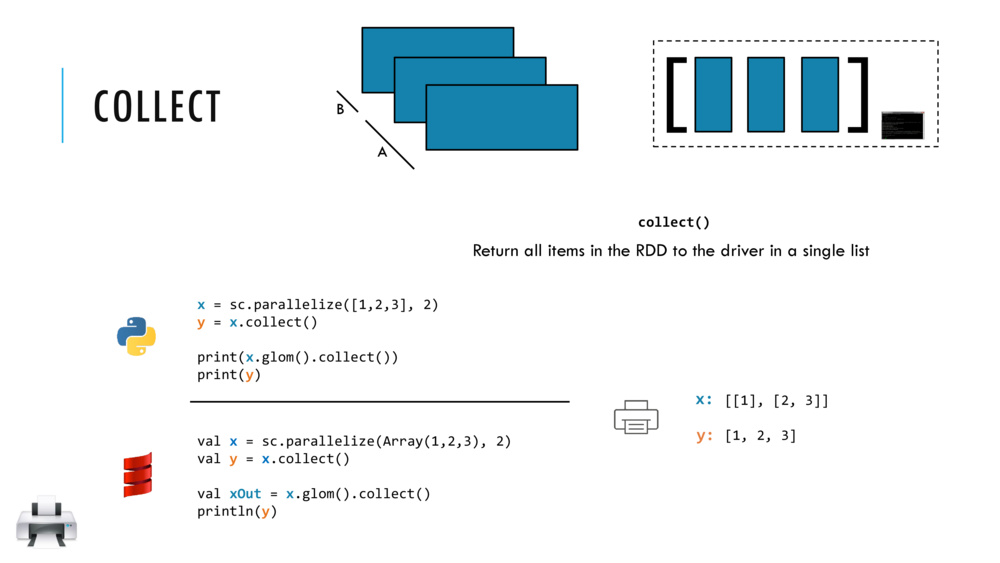
val x = sc.parallelize(Array(1,2,3), 2) // make a RDD with two partitions
x: org.apache.spark.rdd.RDD[Int] = ParallelCollectionRDD[89108] at parallelize at <console>:34
// simply returns all elements in RDD x to the driver as an Array
val y = x.collect()
y: Array[Int] = Array(1, 2, 3)
//glom() flattens elements on the same partition
val xOut = x.glom().collect()
xOut: Array[Array[Int]] = Array(Array(1), Array(2, 3))
getNumPartitions
Return the number of partitions in RDD.
Let us look at the legend and overview of the visual RDD Api.
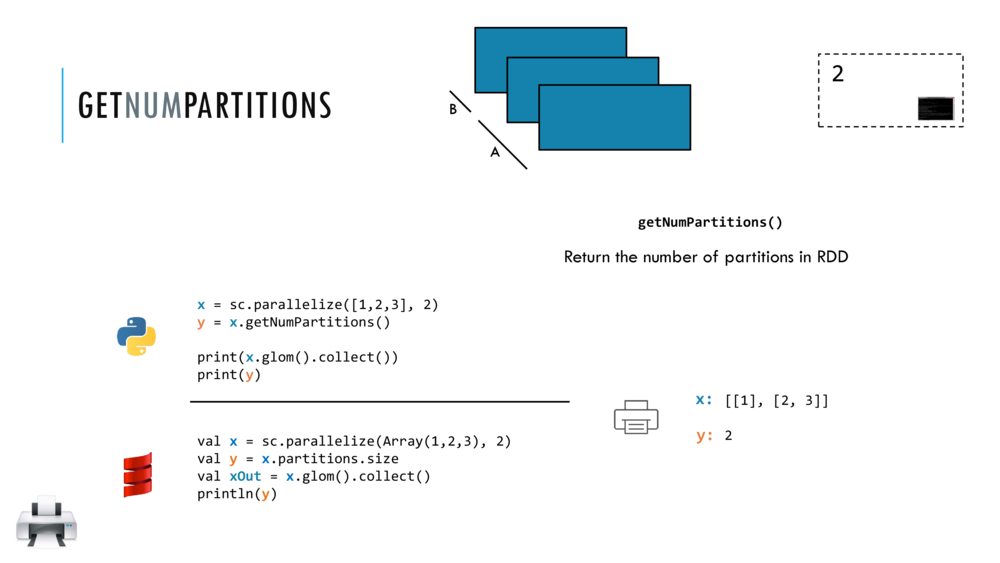
val x = sc.parallelize(Array(1,2,3), 2) // RDD with 2 partitions
x: org.apache.spark.rdd.RDD[Int] = ParallelCollectionRDD[655] at parallelize at <console>:34
val y = x.partitions.size
y: Int = 2
//glom() flattens elements on the same partition
val xOut = x.glom().collect()
xOut: Array[Array[Int]] = Array(Array(1), Array(2, 3))
reduce
Aggregate all the elements of the RDD by applying a user function pairwise to elements and partial results, and returns a result to the driver.
Let us look at the legend and overview of the visual RDD Api.
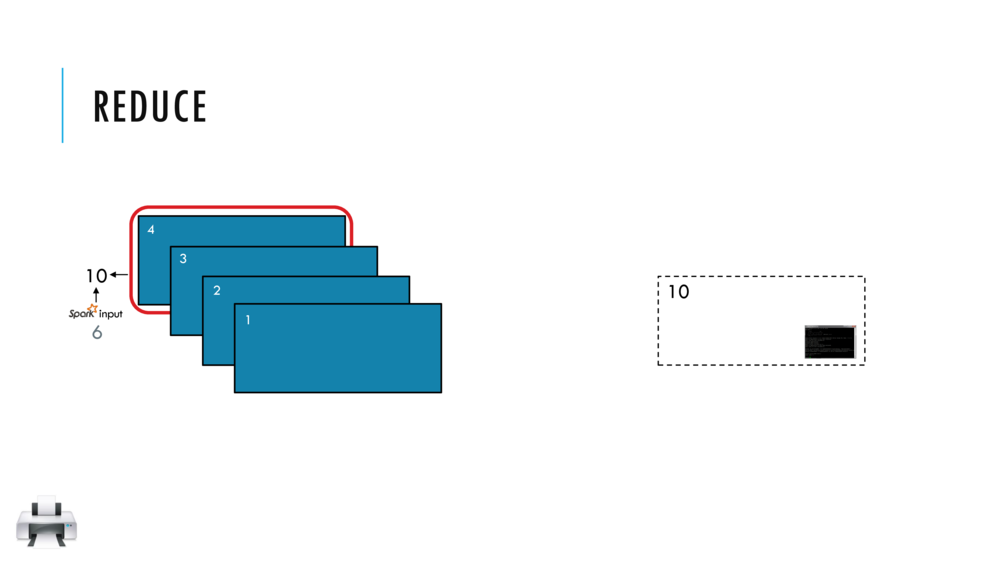
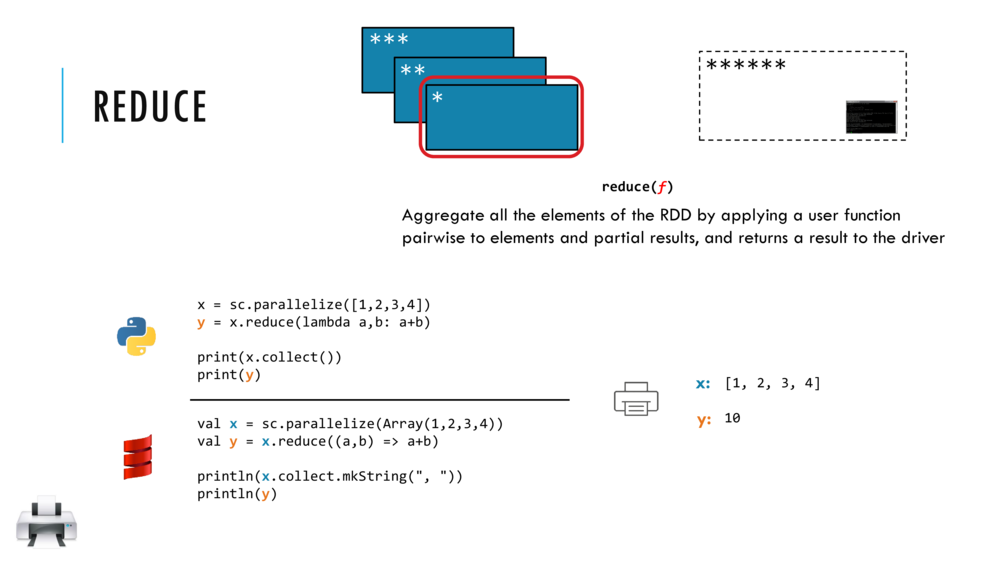
val x = sc.parallelize(Array(1,2,3,4))
val y = x.reduce((a,b) => a+b)
x: org.apache.spark.rdd.RDD[Int] = ParallelCollectionRDD[944] at parallelize at <console>:34
y: Int = 10
println(x.collect.mkString(", "))
println(y)
1, 2, 3, 4
10
coalesce
Return a new RDD which is reduced to a smaller number of partitions.
Let us look at the legend and overview of the visual RDD Api.
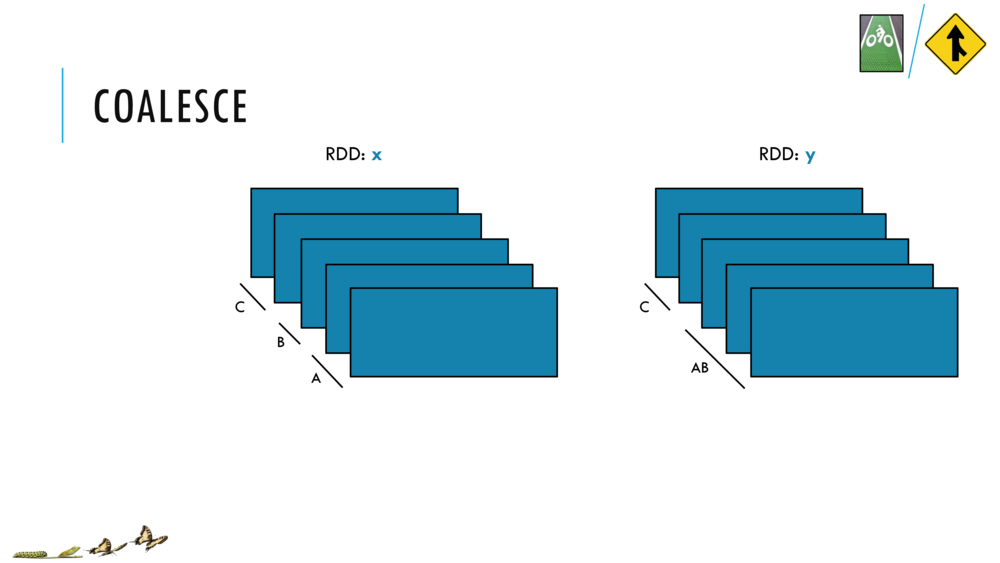
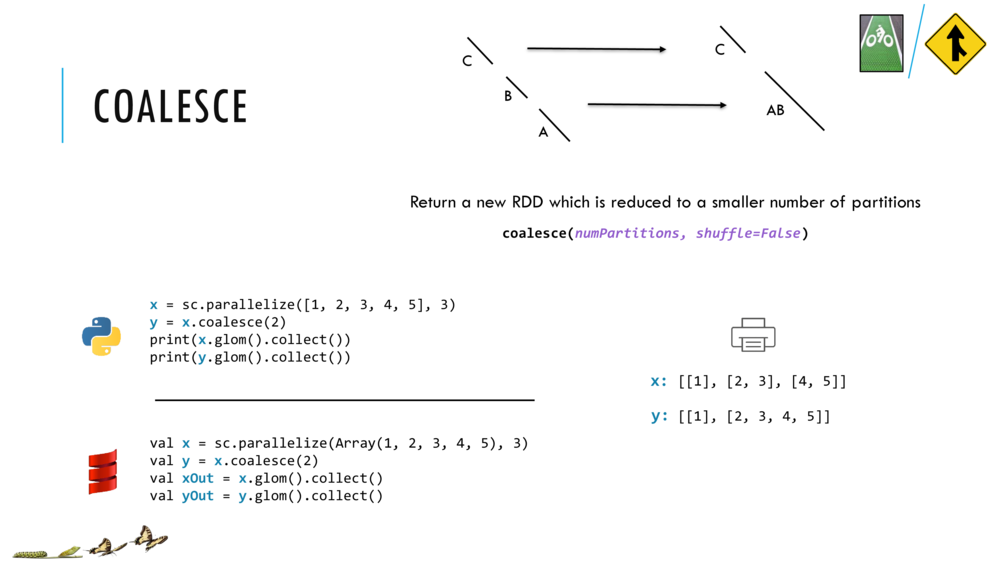
val x = sc.parallelize(Array(1,2,3,4,5), 3) // make RDD with 3 partitions
x: org.apache.spark.rdd.RDD[Int] = ParallelCollectionRDD[1419] at parallelize at <console>:34
x.collect()
res0: Array[Int] = Array(1, 2, 3, 4, 5)
val y = x.coalesce(2) // coalesce RDD x from 3 to 2 partitions
y: org.apache.spark.rdd.RDD[Int] = CoalescedRDD[1428] at coalesce at <console>:36
y.collect() // x and y will yield the same list to driver upon collect
res1: Array[Int] = Array(1, 2, 3, 4, 5)
//glom() flattens elements on the same partition
val xOut = x.glom().collect() // x has three partitions
xOut: Array[Array[Int]] = Array(Array(1), Array(2, 3), Array(4, 5))
val yOut = y.glom().collect() // However y has only 2 partitions
yOut: Array[Array[Int]] = Array(Array(1), Array(2, 3, 4, 5))
distinct
Return a new RDD containing distinct items from the original RDD (omitting all duplicates).
Let us look at the legend and overview of the visual RDD Api.
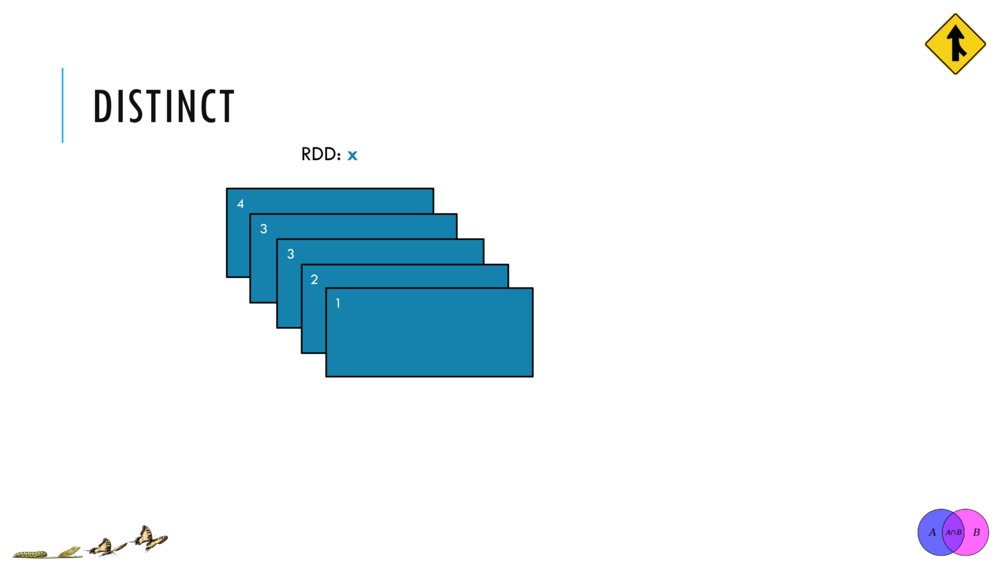
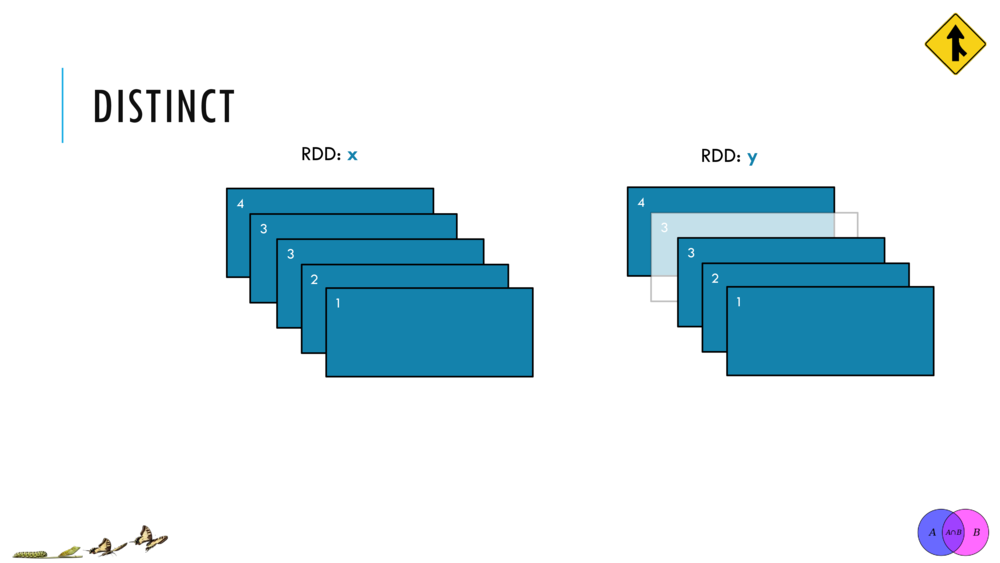
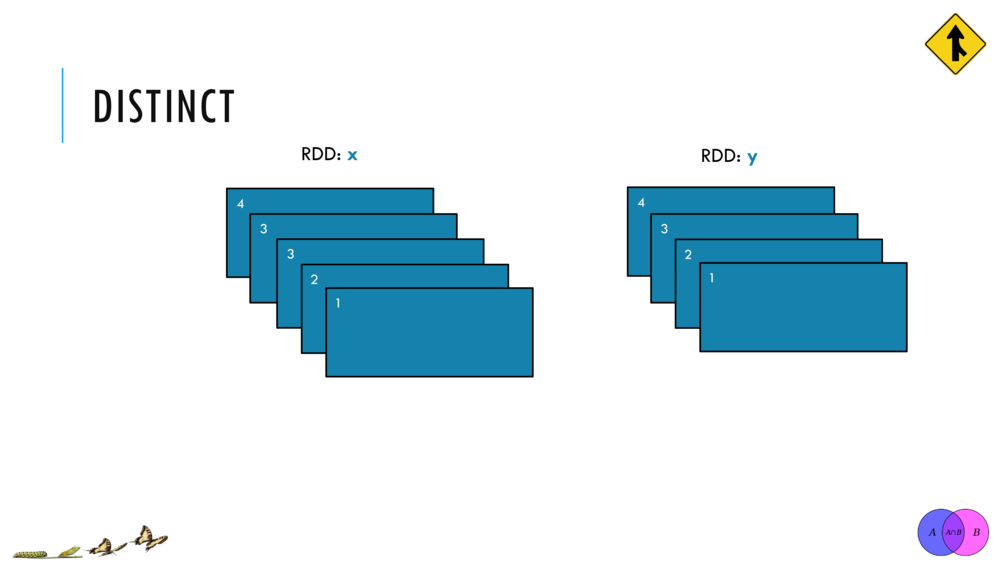
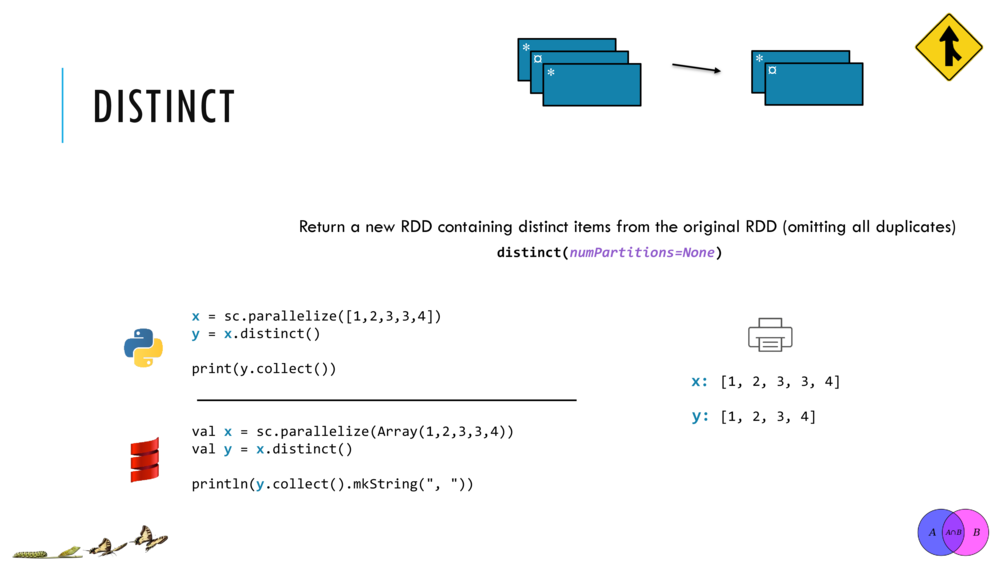
val x = sc.parallelize(Array(1,2,3,3,4))
val y = x.distinct()
x: org.apache.spark.rdd.RDD[Int] = ParallelCollectionRDD[1320] at parallelize at <console>:36
y: org.apache.spark.rdd.RDD[Int] = MapPartitionsRDD[1323] at distinct at <console>:37
y.collect()
res2: Array[Int] = Array(4, 2, 1, 3)
filter
Return a new RDD containing only elements of this RDD that satisfy a predicate.
Let us look at the legend and overview of the visual RDD Api.
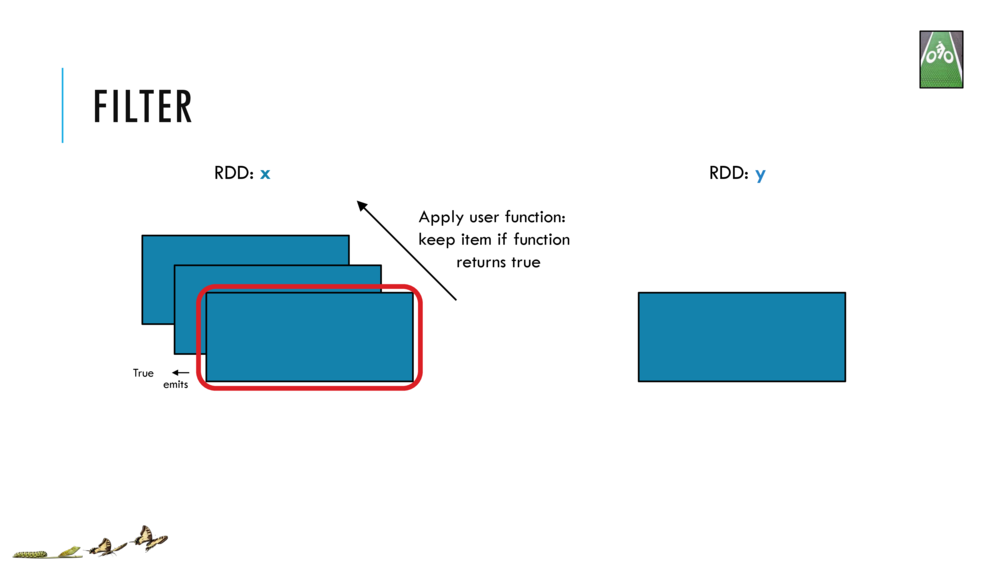
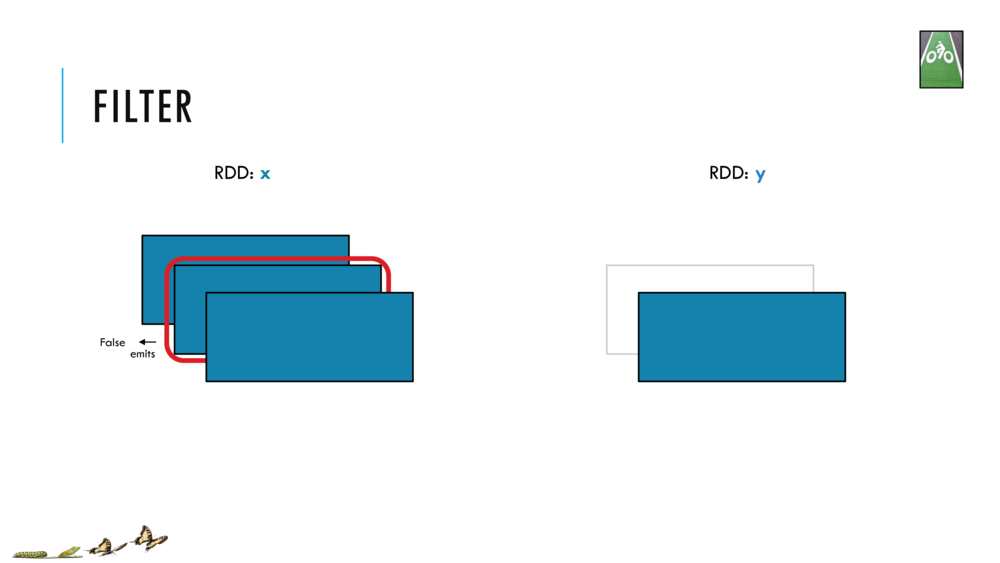
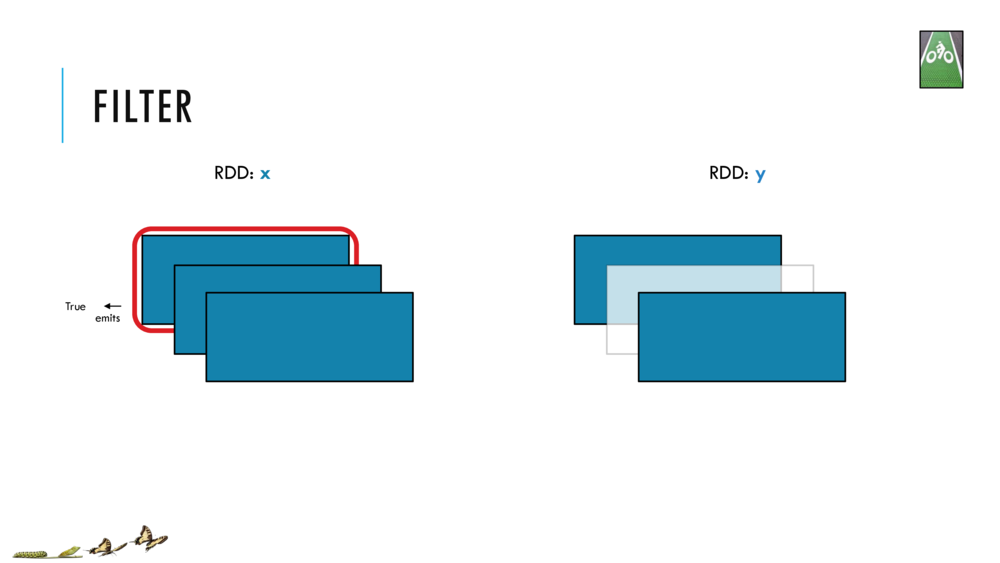
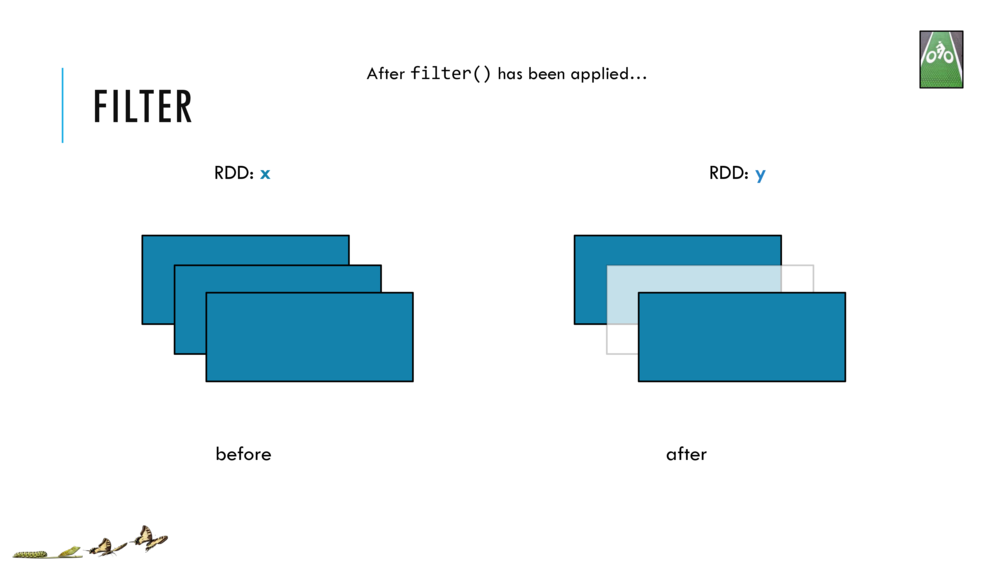
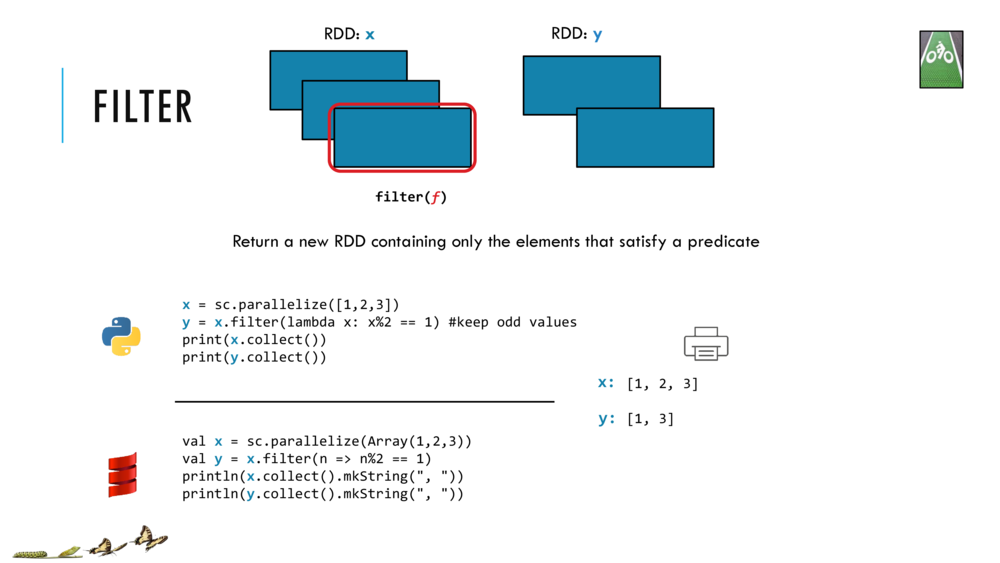
val x = sc.parallelize(Array(1,2,3))
val y = x.filter(n => n%2 == 1)
x: org.apache.spark.rdd.RDD[Int] = ParallelCollectionRDD[374] at parallelize at <console>:34
y: org.apache.spark.rdd.RDD[Int] = MapPartitionsRDD[375] at filter at <console>:35
println(x.collect().mkString(", "))
println(y.collect().mkString(", "))
1, 2, 3
1, 3
flatMap
Return a new RDD by first applying a function to each element of this RDD and then flattening the results.
Let us look at the legend and overview of the visual RDD Api.
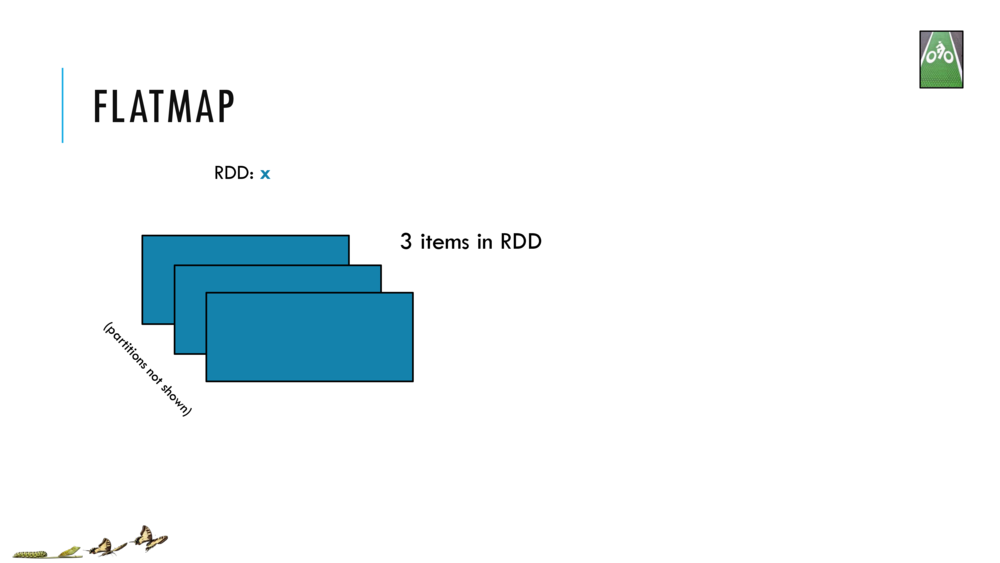
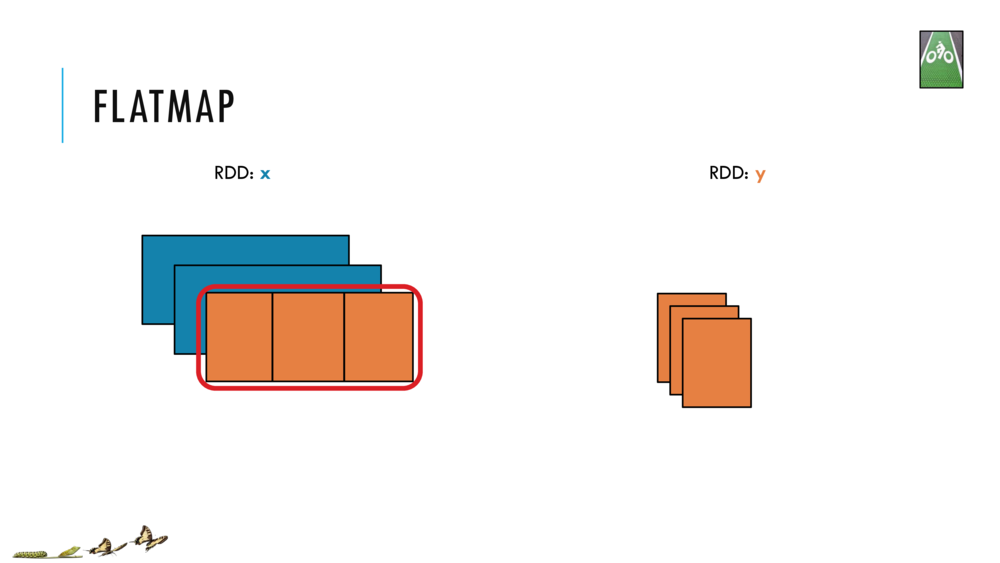
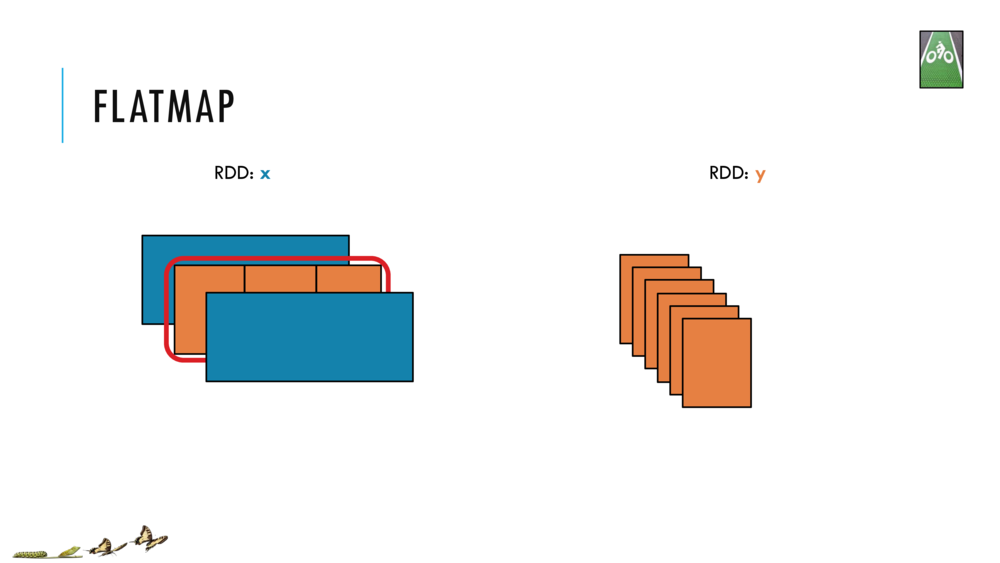
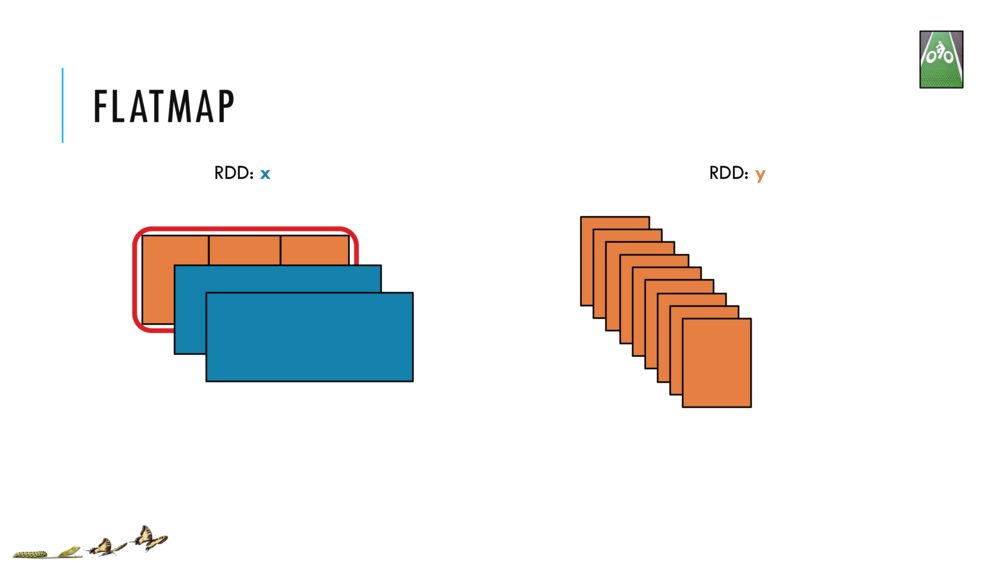
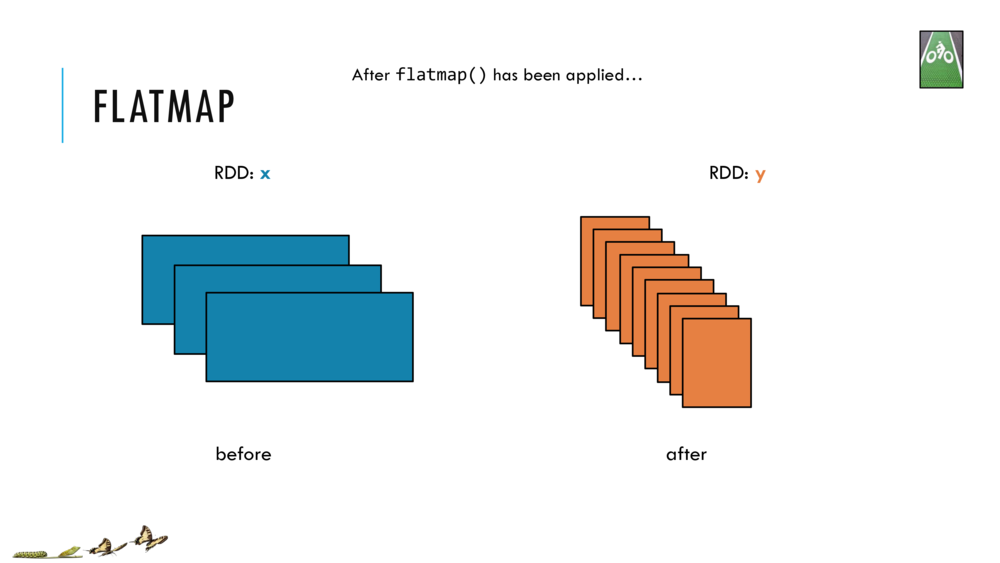
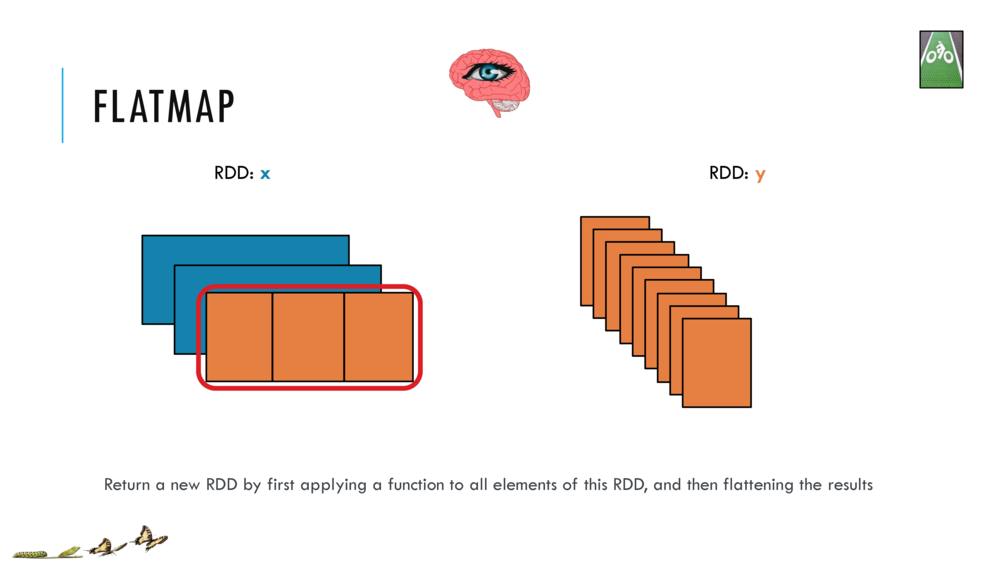
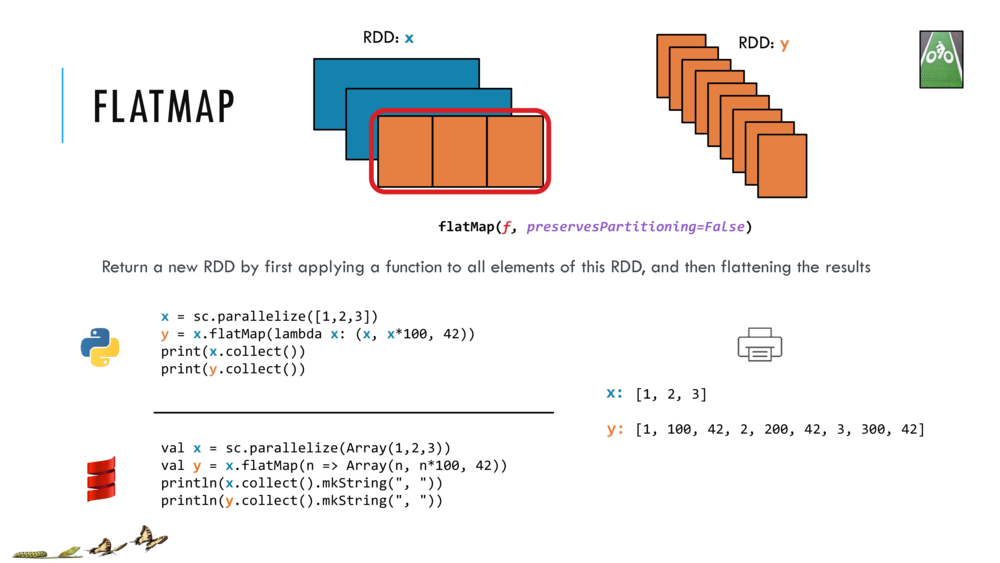
val x = sc.parallelize(Array(1,2,3))
val y = x.flatMap(n => Array(n, n*100, 42))
x: org.apache.spark.rdd.RDD[Int] = ParallelCollectionRDD[289] at parallelize at <console>:34
y: org.apache.spark.rdd.RDD[Int] = MapPartitionsRDD[290] at flatMap at <console>:35
println(x.collect().mkString(", "))
println(y.collect().mkString(", "))
1, 2, 3
1, 100, 42, 2, 200, 42, 3, 300, 42
groupByKey
Group the values for each key in the original RDD. Create a new pair where the original key corresponds to this collected group of values.
Let us look at the legend and overview of the visual RDD Api.
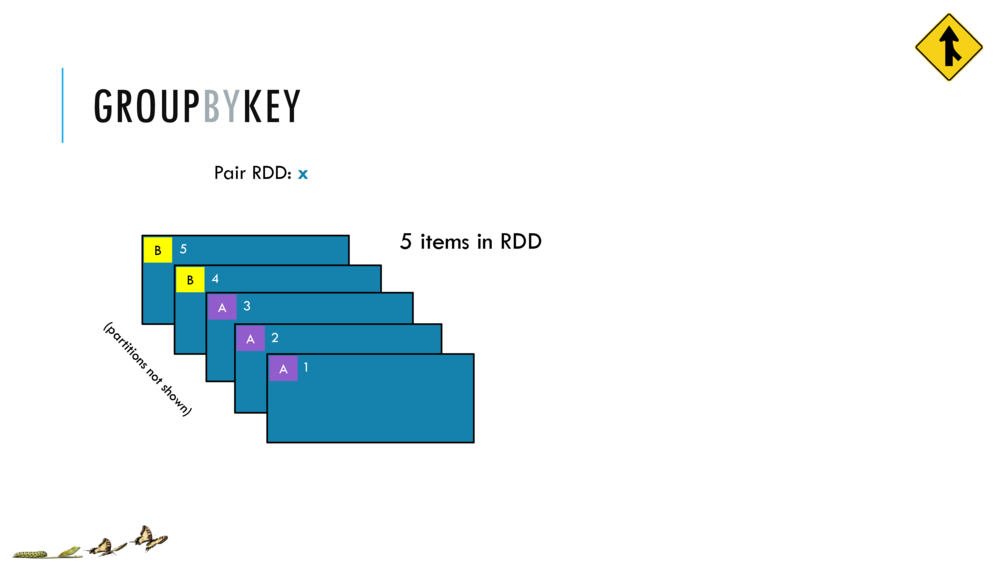
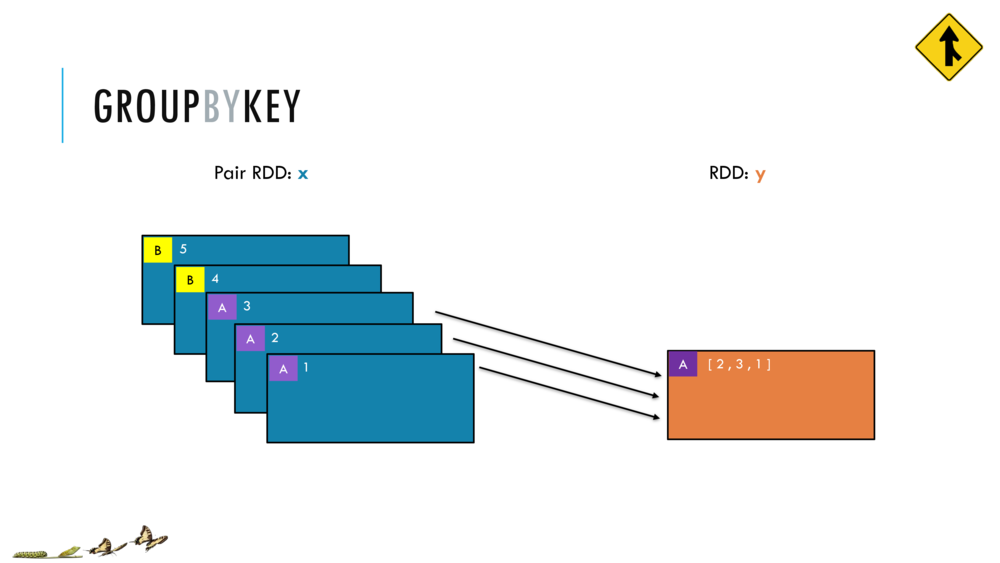
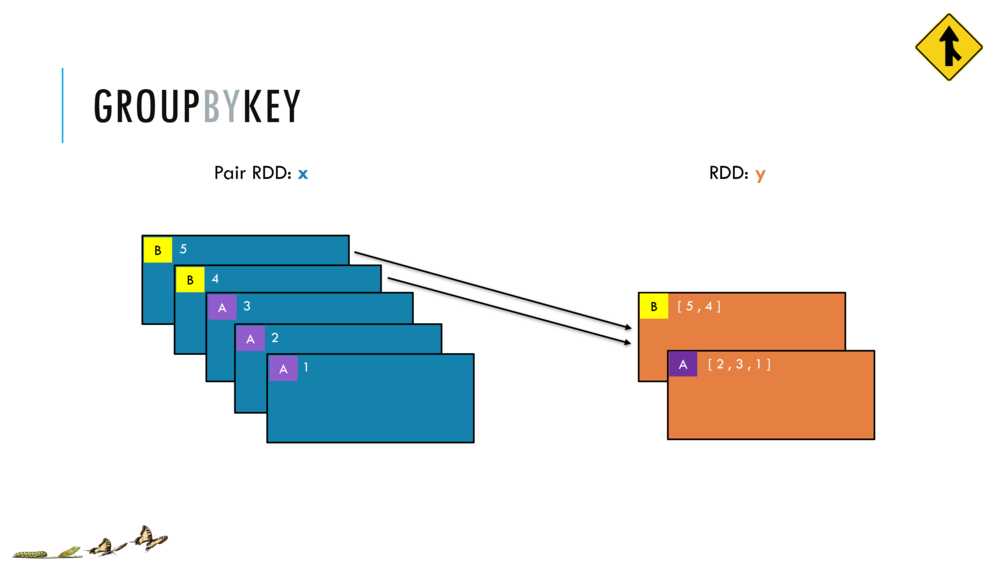
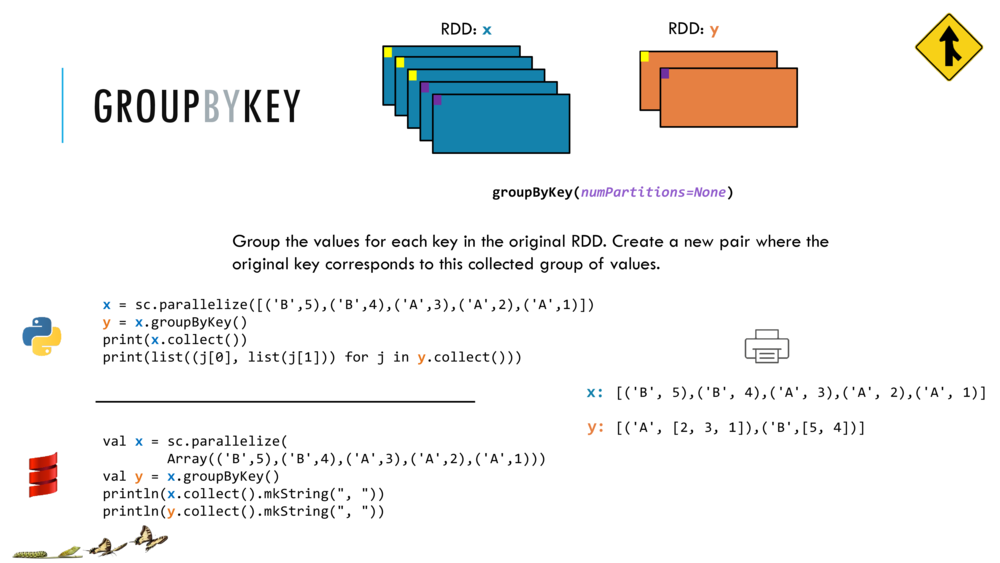
val x = sc.parallelize(Array(('B',5),('B',4),('A',3),('A',2),('A',1)))
val y = x.groupByKey()
x: org.apache.spark.rdd.RDD[(Char, Int)] = ParallelCollectionRDD[476] at parallelize at <console>:34
y: org.apache.spark.rdd.RDD[(Char, Iterable[Int])] = ShuffledRDD[477] at groupByKey at <console>:35
println(x.collect().mkString(", "))
println(y.collect().mkString(", "))
(B,5), (B,4), (A,3), (A,2), (A,1)
(B,CompactBuffer(5, 4)), (A,CompactBuffer(3, 2, 1))
groupBy
Group the data in the original RDD. Create pairs where the key is the output of a user function, and the value is all items for which the function yield this key.
Let us look at the legend and overview of the visual RDD Api.
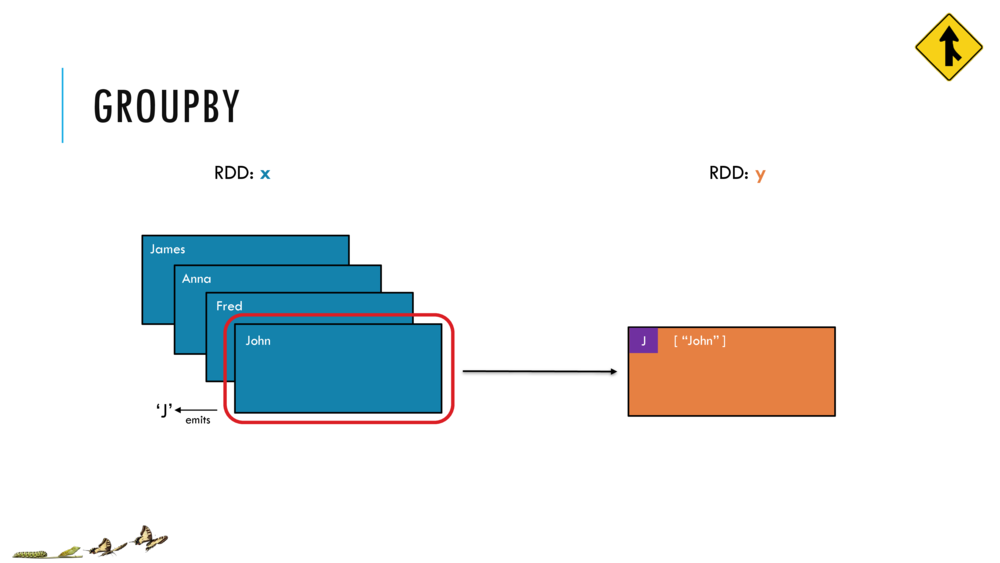
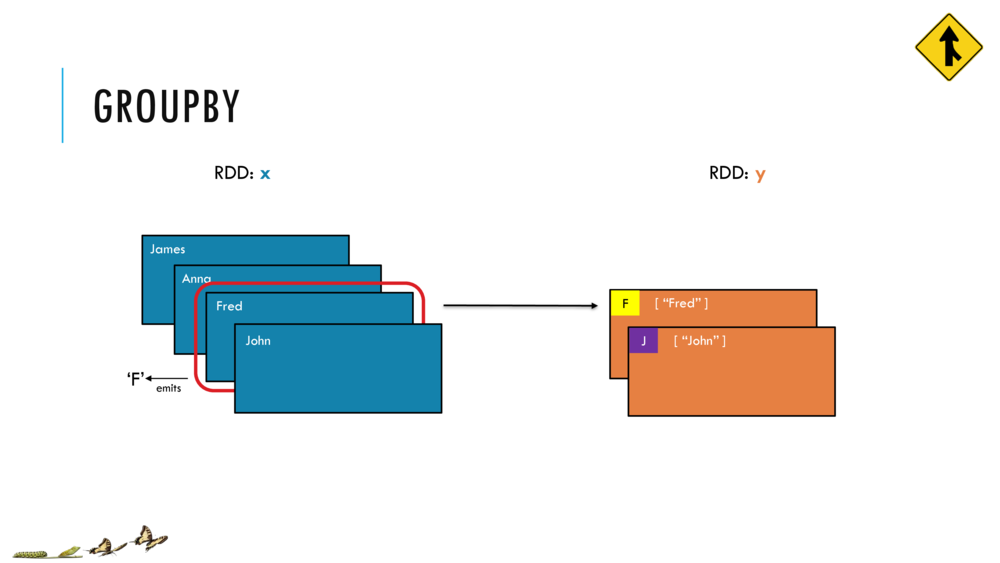
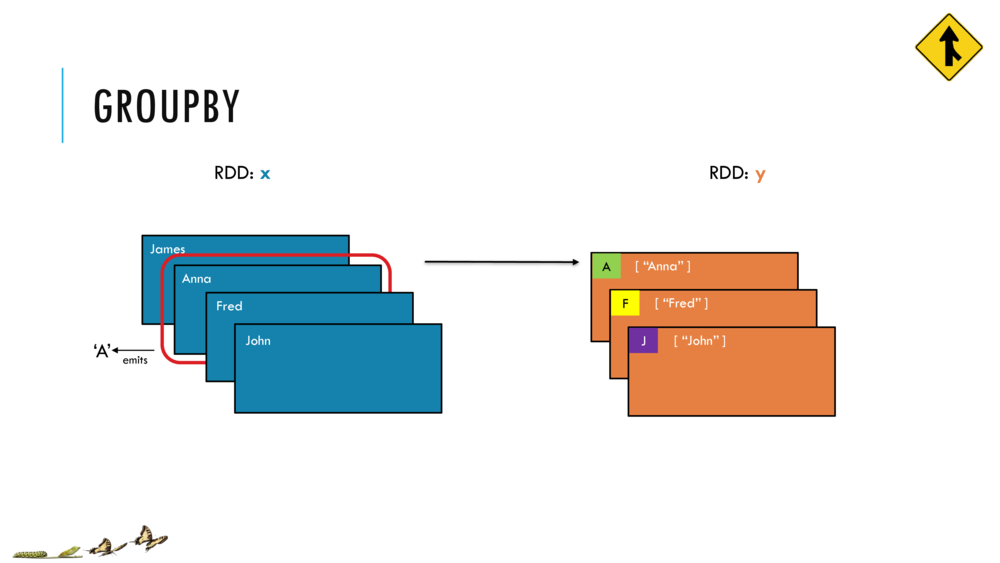
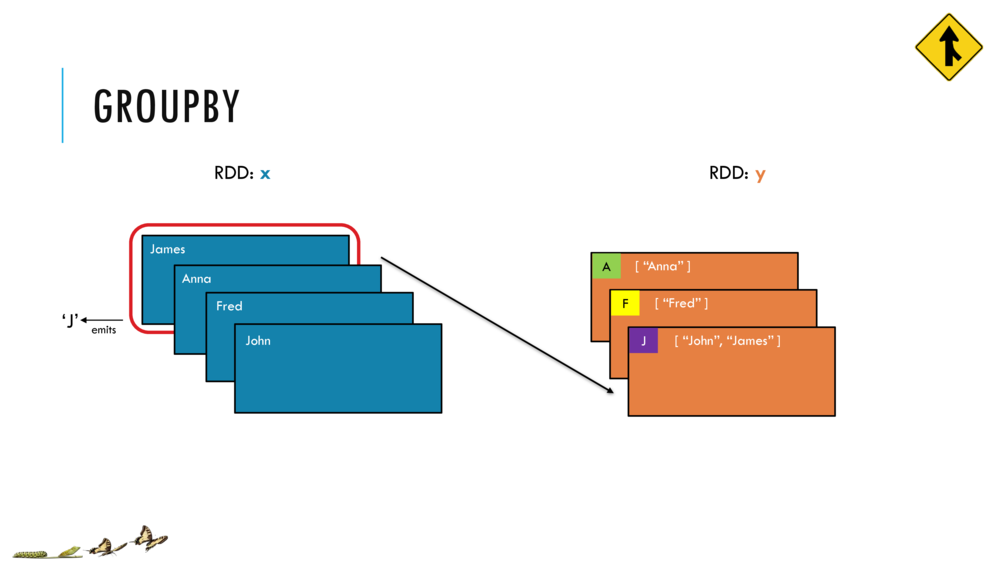
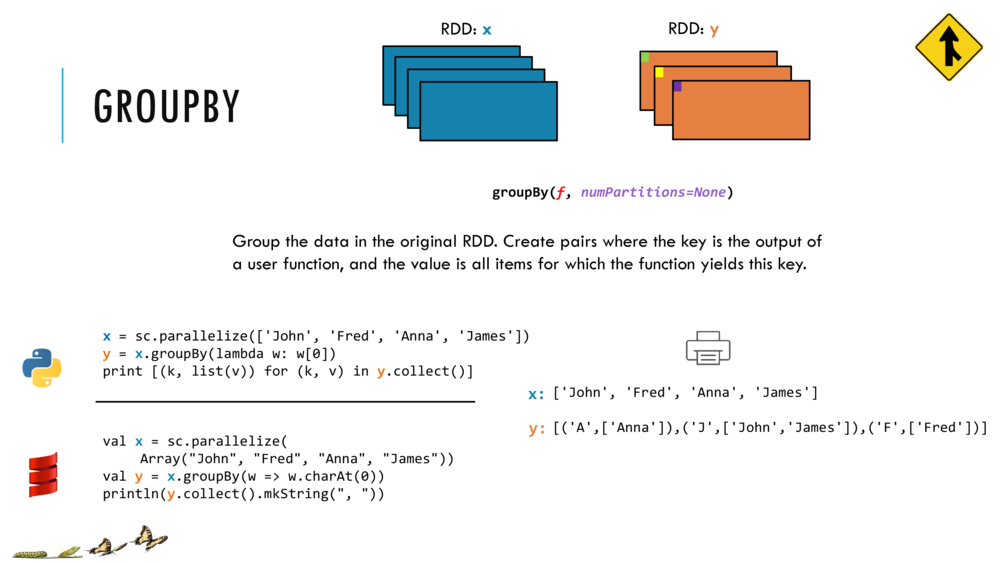
val x = sc.parallelize(Array("John", "Fred", "Anna", "James"))
val y = x.groupBy(w => w.charAt(0))
x: org.apache.spark.rdd.RDD[String] = ParallelCollectionRDD[366] at parallelize at <console>:34
y: org.apache.spark.rdd.RDD[(Char, Iterable[String])] = ShuffledRDD[368] at groupBy at <console>:35
println(x.collect().mkString(", "))
println(y.collect().mkString(", "))
John, Fred, Anna, James
(J,CompactBuffer(John, James)), (F,CompactBuffer(Fred)), (A,CompactBuffer(Anna))
join
Return a new RDD containing all pairs of elements having the same key in the original RDDs.
Let us look at the legend and overview of the visual RDD Api.
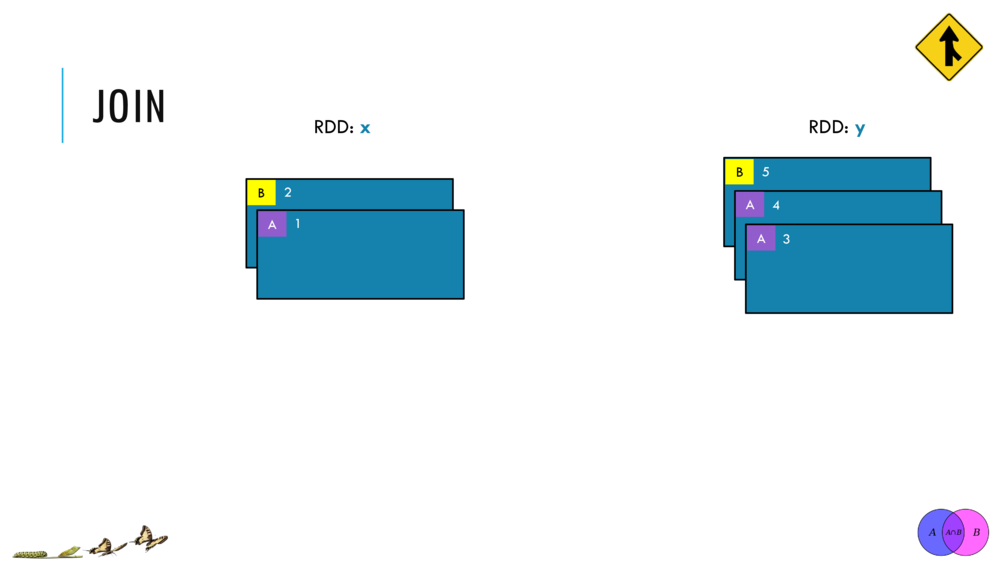
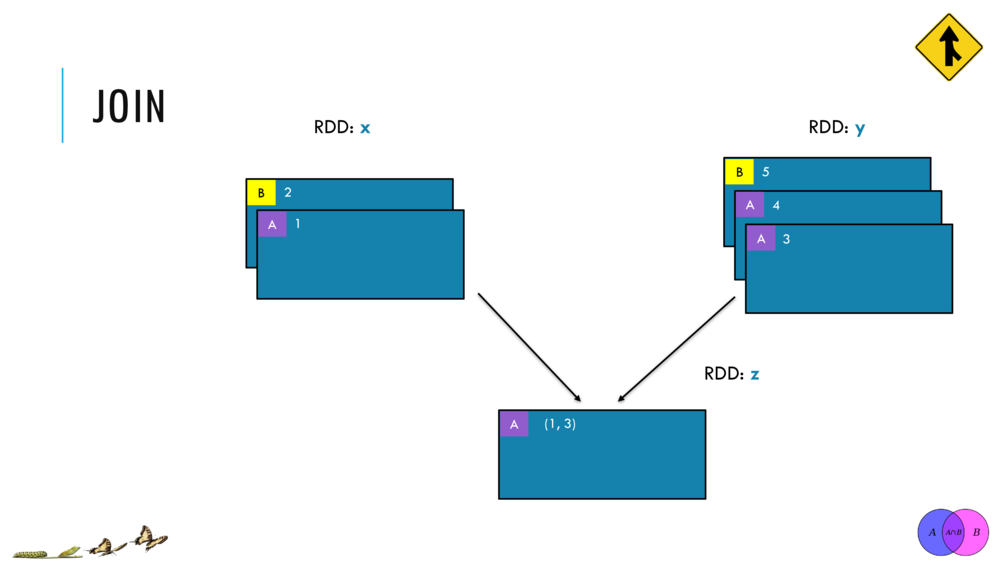
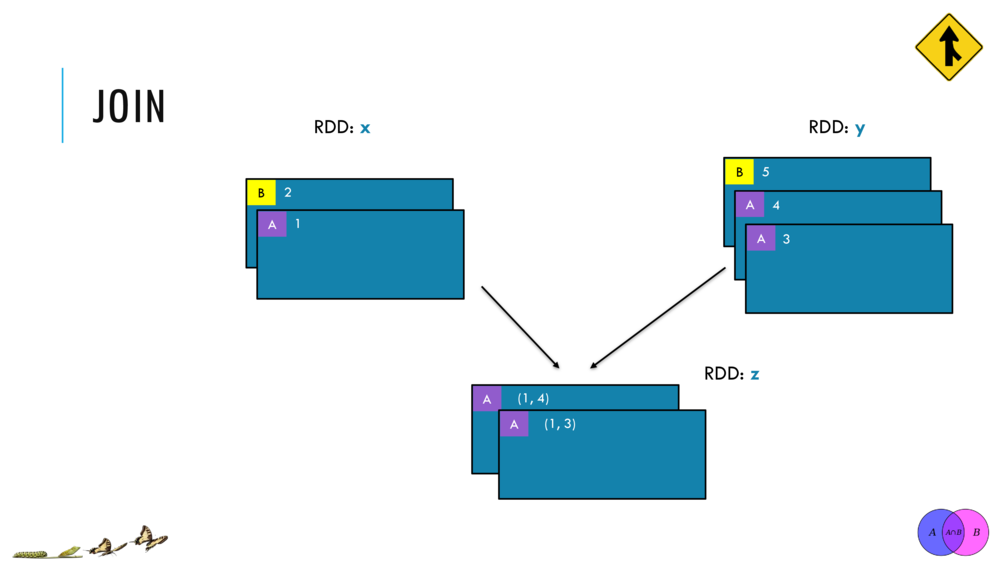
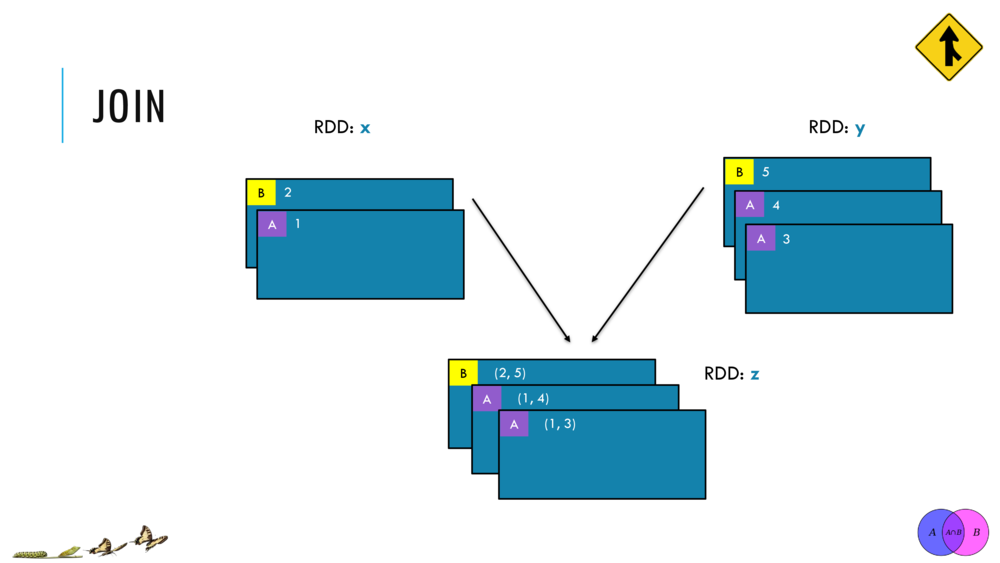
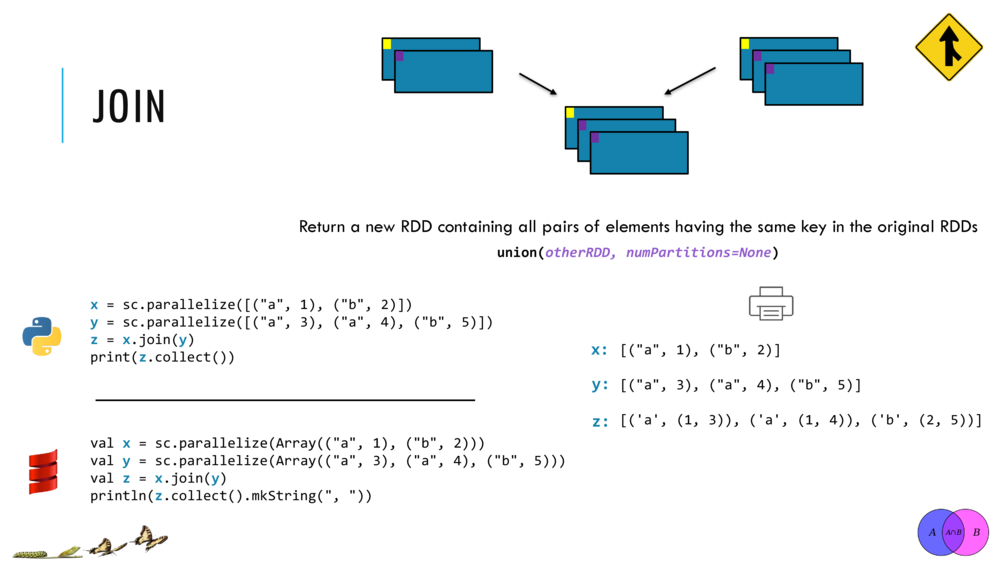
val x = sc.parallelize(Array(("a", 1), ("b", 2)))
val y = sc.parallelize(Array(("a", 3), ("a", 4), ("b", 5)))
x: org.apache.spark.rdd.RDD[(String, Int)] = ParallelCollectionRDD[1159] at parallelize at <console>:34
y: org.apache.spark.rdd.RDD[(String, Int)] = ParallelCollectionRDD[1160] at parallelize at <console>:35
val z = x.join(y)
z: org.apache.spark.rdd.RDD[(String, (Int, Int))] = MapPartitionsRDD[1178] at join at <console>:38
z.collect()
res2: Array[(String, (Int, Int))] = Array((b,(2,5)), (a,(1,3)), (a,(1,4)))
keyBy
Create a Pair RDD, forming one pair for each item in the original RDD. The pair's key is calculated from the value via a user-supplied function.
Let us look at the legend and overview of the visual RDD Api.
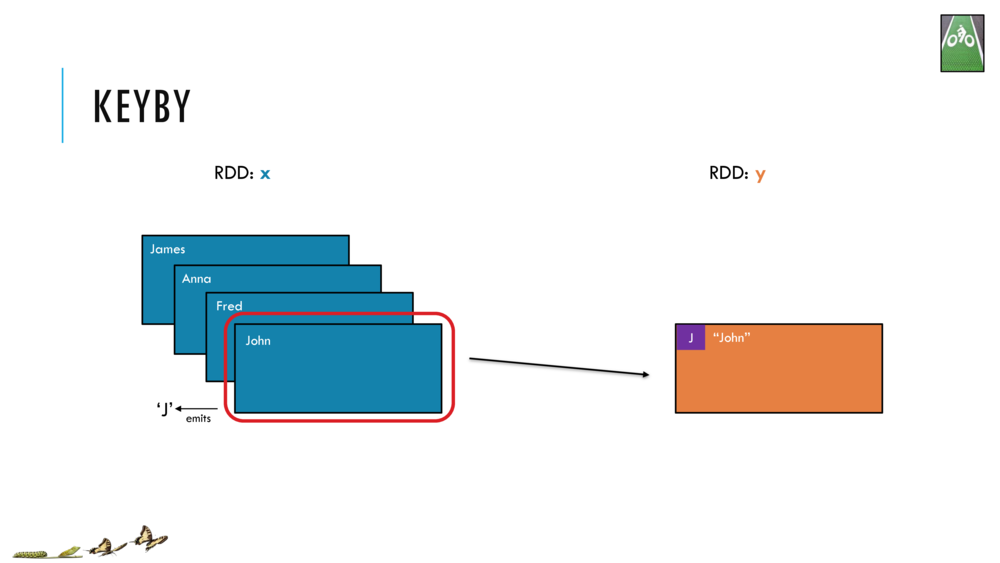
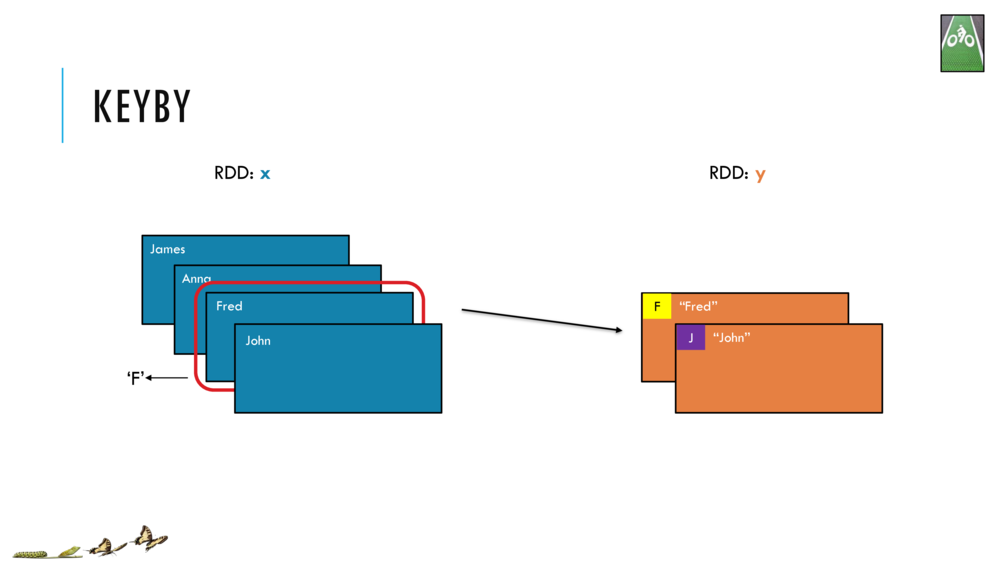
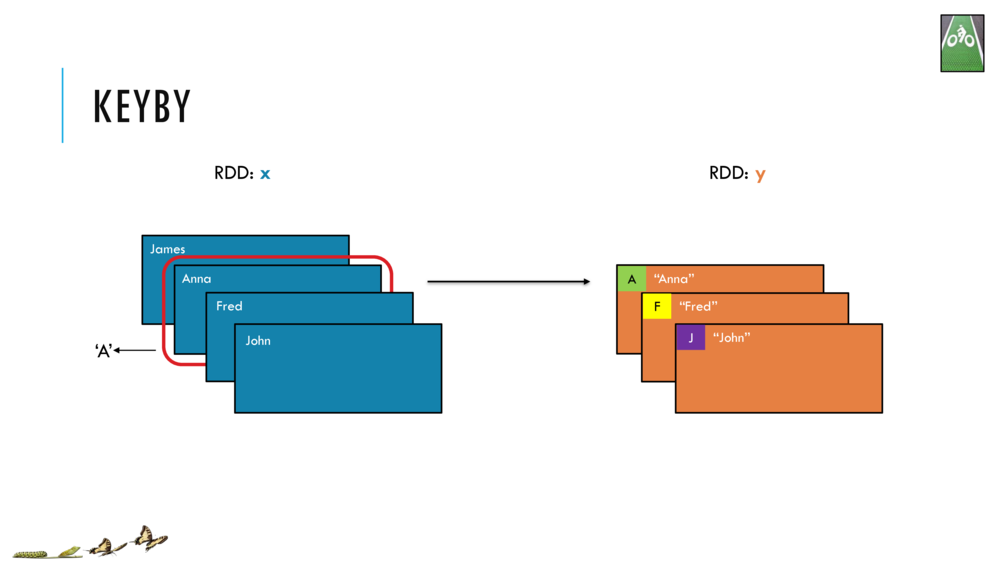
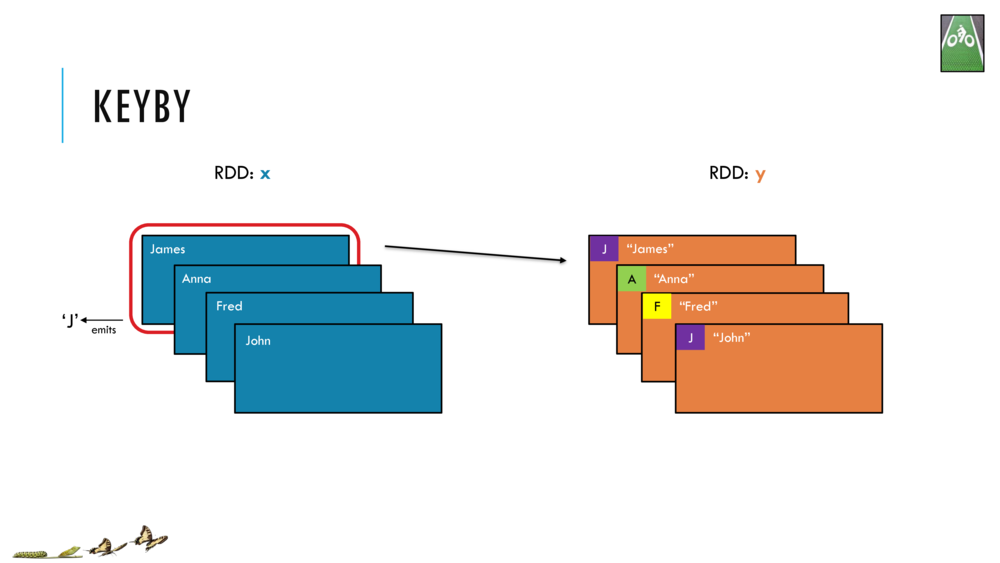
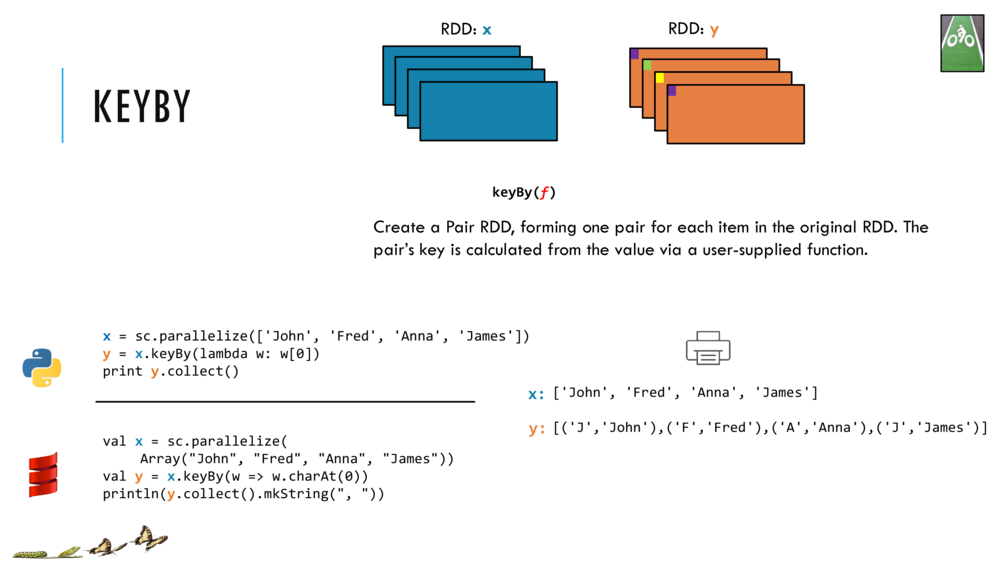
val x = sc.parallelize(Array("John", "Fred", "Anna", "James"))
val y = x.keyBy(w => w.charAt(0))
println(y.collect().mkString(", "))
map
Return a new RDD by applying a function to each element of this RDD.
Let us look at the legend and overview of the visual RDD Api.
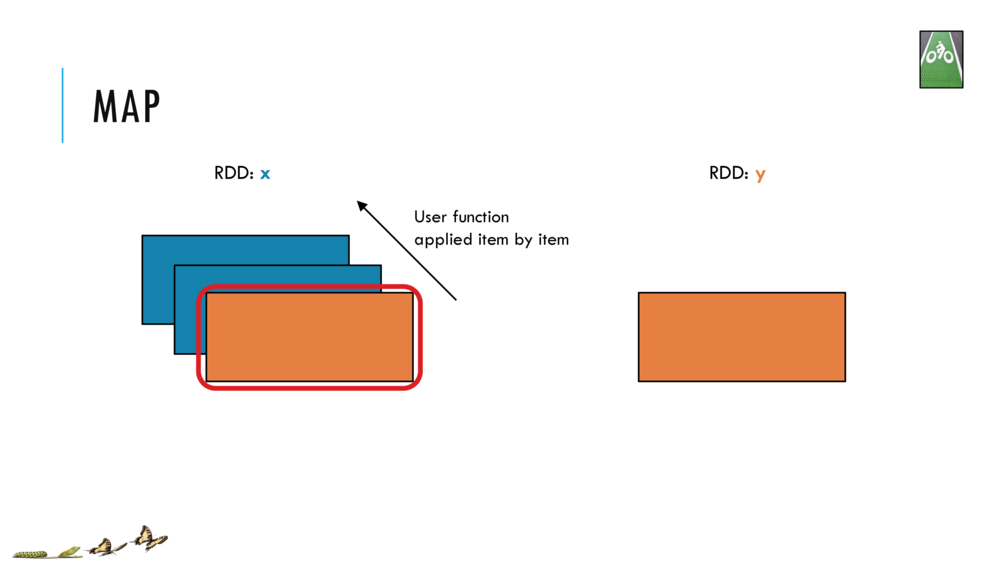
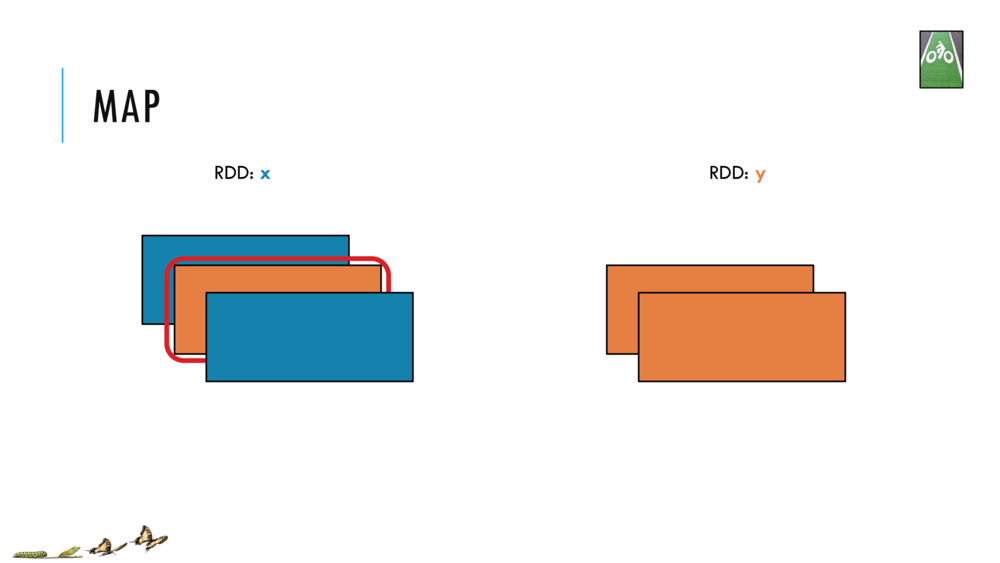
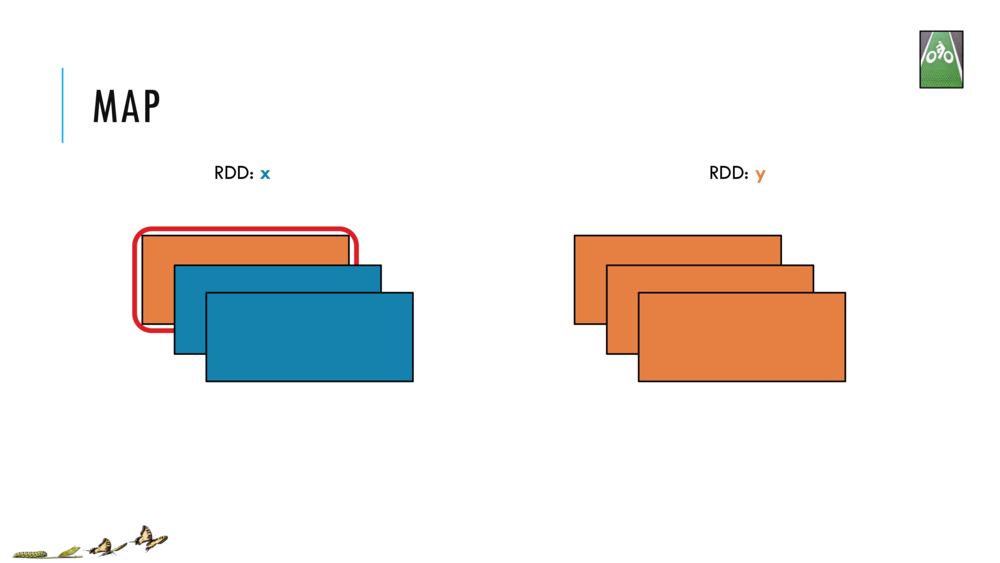
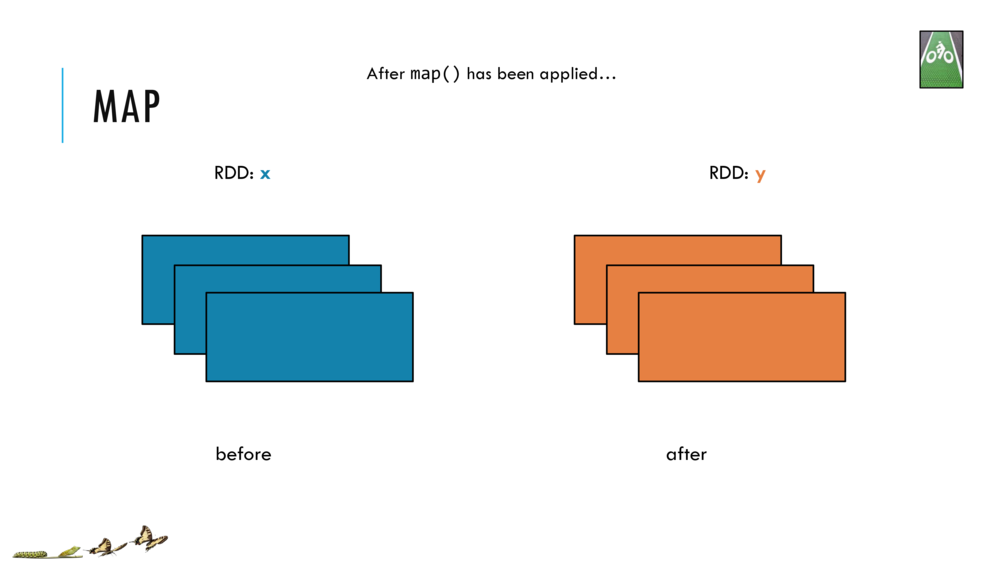
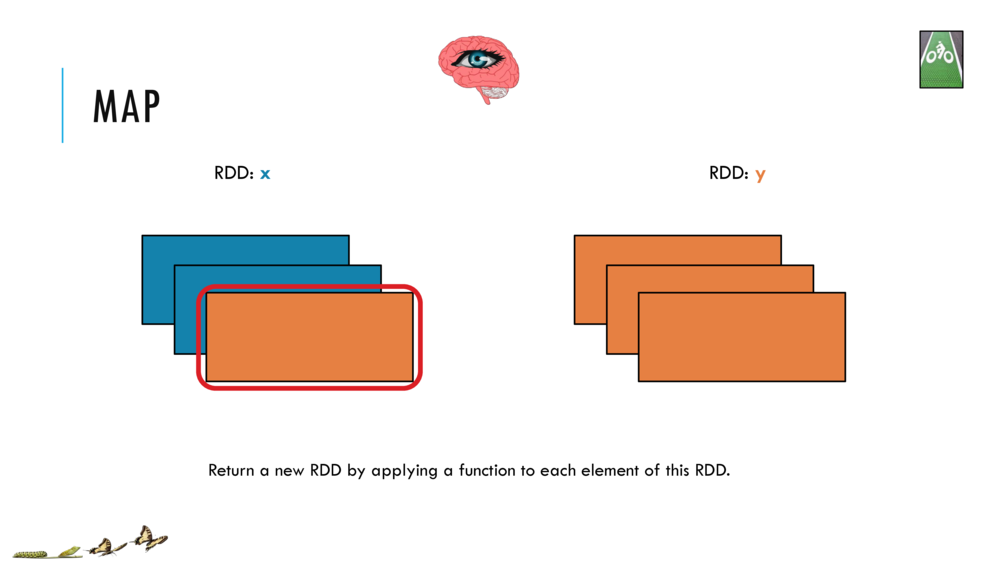
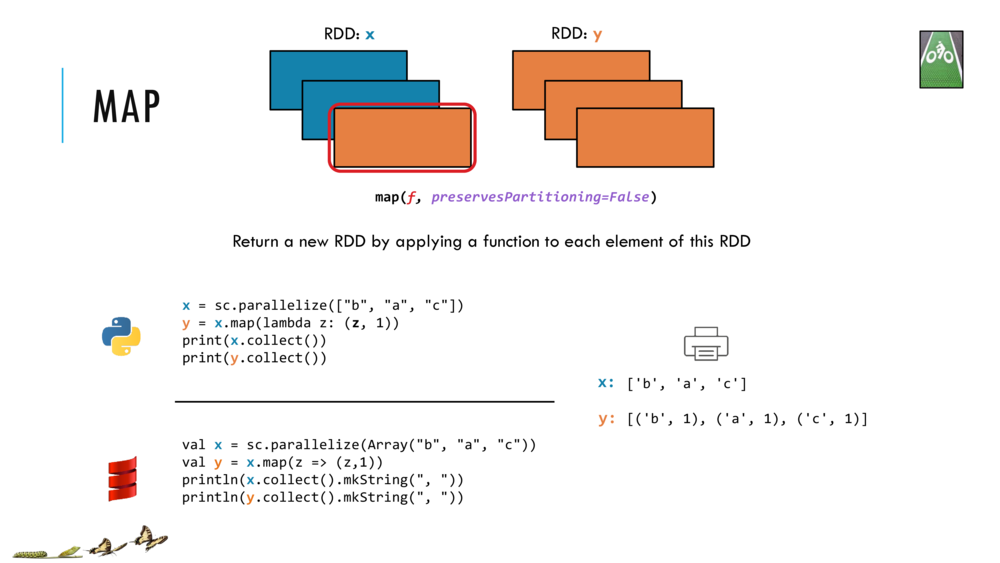
val x = sc.parallelize(Array("b", "a", "c"))
val y = x.map(z => (z,1))
x: org.apache.spark.rdd.RDD[String] = ParallelCollectionRDD[176] at parallelize at <console>:36
y: org.apache.spark.rdd.RDD[(String, Int)] = MapPartitionsRDD[177] at map at <console>:37
println(x.collect().mkString(","))
println(y.collect().mkString(","))
b,a,c
(b,1),(a,1),(c,1)
mapPartitions
Return a new RDD by applying a function to each partition of the RDD.
Let us look at the legend and overview of the visual RDD Api.
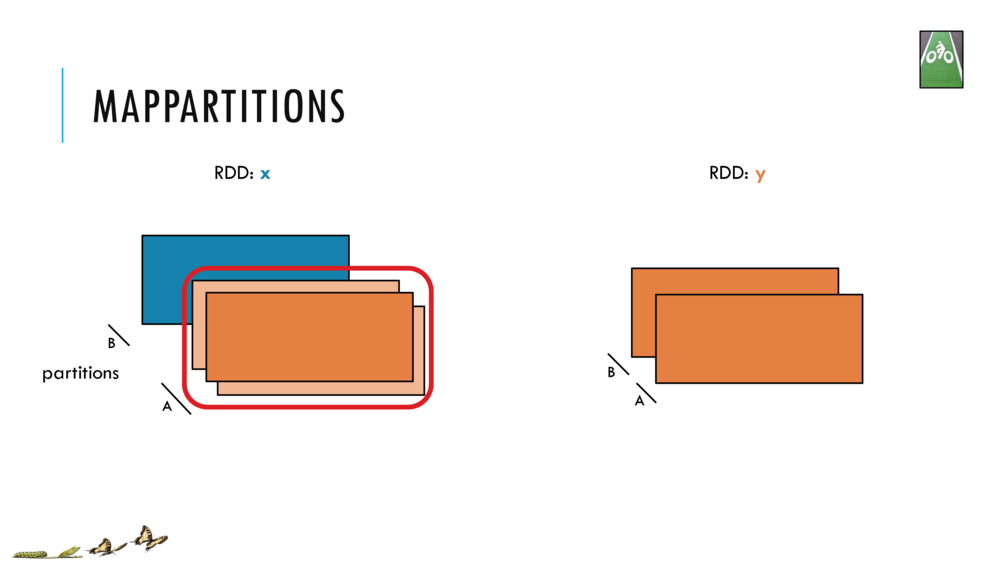
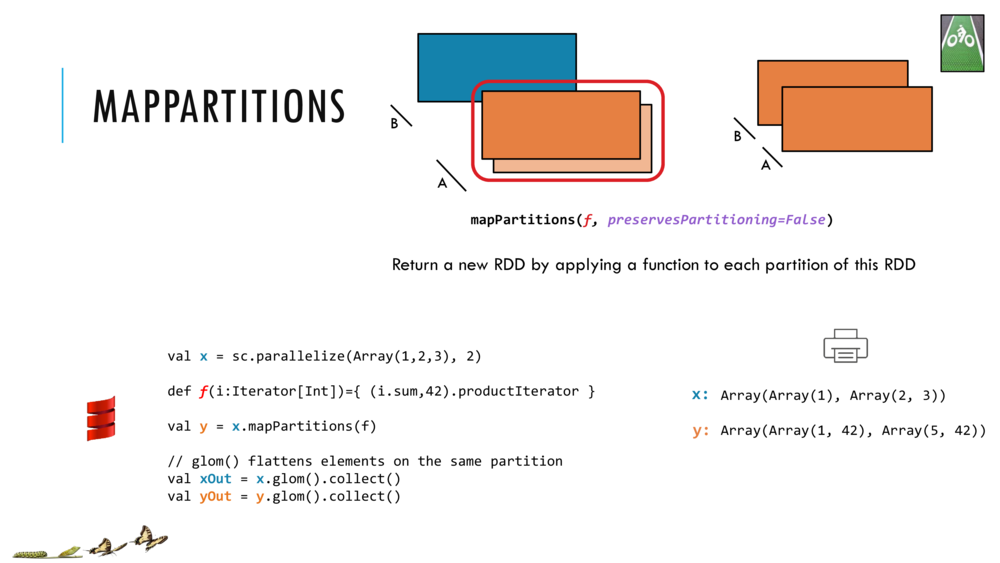
val x = sc.parallelize(Array(1,2,3), 2) // RDD with 2 partitions
x: org.apache.spark.rdd.RDD[Int] = ParallelCollectionRDD[10] at parallelize at <console>:34
def f(i:Iterator[Int])={ (i.sum, 42).productIterator }
f: (i: Iterator[Int])Iterator[Any]
val y = x.mapPartitions(f)
y: org.apache.spark.rdd.RDD[Any] = MapPartitionsRDD[12] at mapPartitions at <console>:38
// glom() flattens elements on the same partition
val xOut = x.glom().collect()
xOut: Array[Array[Int]] = Array(Array(1), Array(2, 3))
val yOut = y.glom().collect()
yOut: Array[Array[Any]] = Array(Array(1, 42), Array(5, 42))
mapPartitionsWithIndex
Return a new RDD by applying a function to each partition of this RDD, while tracking the index of the original partition.
Let us look at the legend and overview of the visual RDD Api.
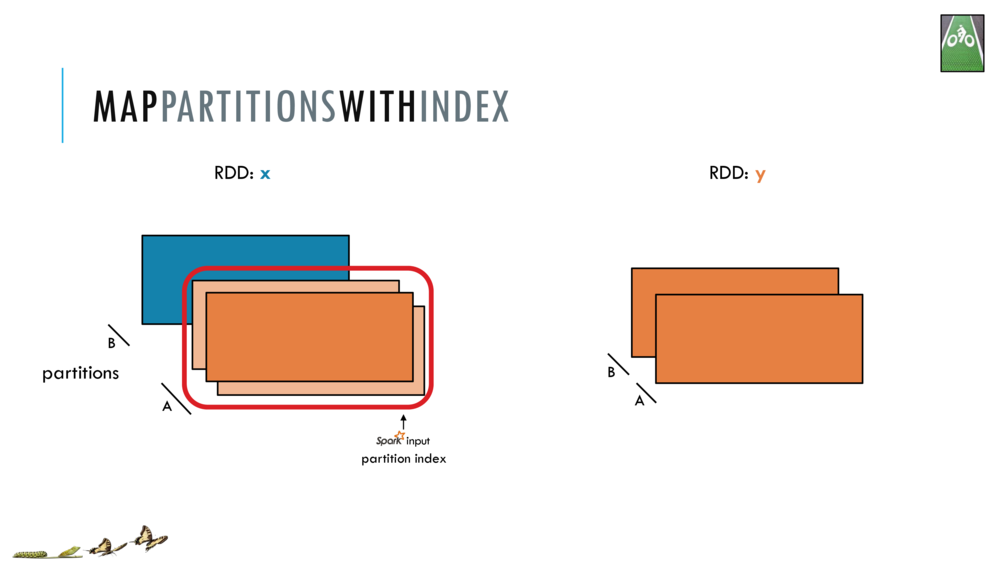
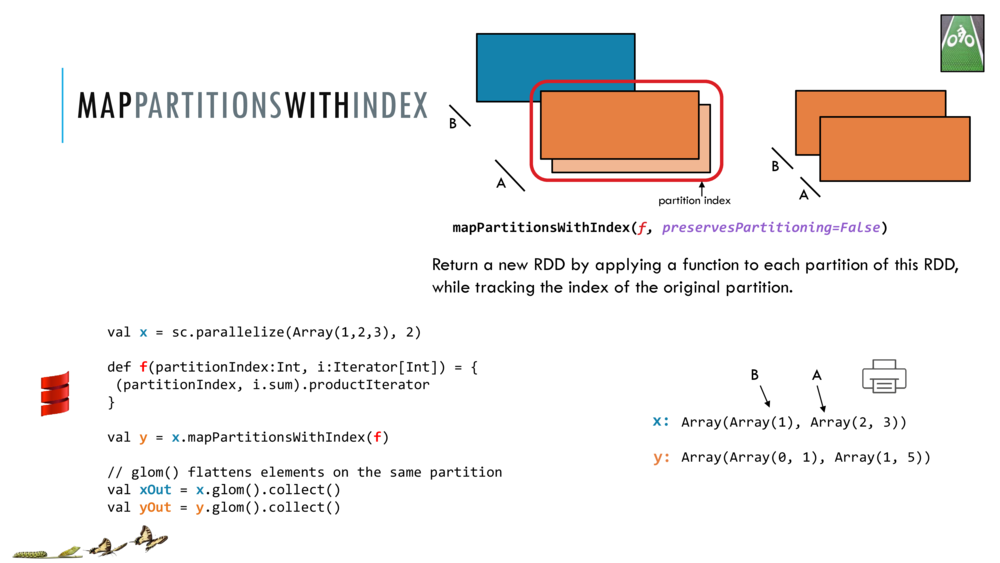
val x = sc. parallelize(Array(1,2,3), 2) // make an RDD with 2 partitions
x: org.apache.spark.rdd.RDD[Int] = ParallelCollectionRDD[289] at parallelize at <console>:34
def f(partitionIndex:Int, i:Iterator[Int]) = {
(partitionIndex, i.sum).productIterator
}
f: (partitionIndex: Int, i: Iterator[Int])Iterator[Any]
val y = x.mapPartitionsWithIndex(f)
y: org.apache.spark.rdd.RDD[Any] = MapPartitionsRDD[304] at mapPartitionsWithIndex at <console>:38
//glom() flattens elements on the same partition
val xout = x.glom().collect()
xout: Array[Array[Int]] = Array(Array(1), Array(2, 3))
val yout = y.glom().collect()
yout: Array[Array[Any]] = Array(Array(0, 1), Array(1, 5))
partitionBy
Return a new RDD with the specified number of partitions, placing original items into the partition returned by a user supplied function.
Let us look at the legend and overview of the visual RDD Api.
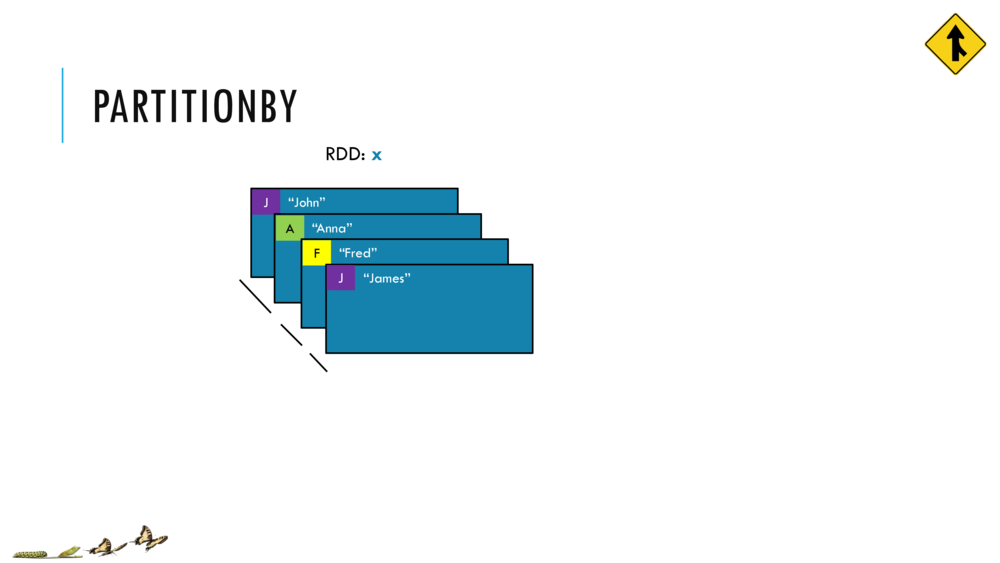
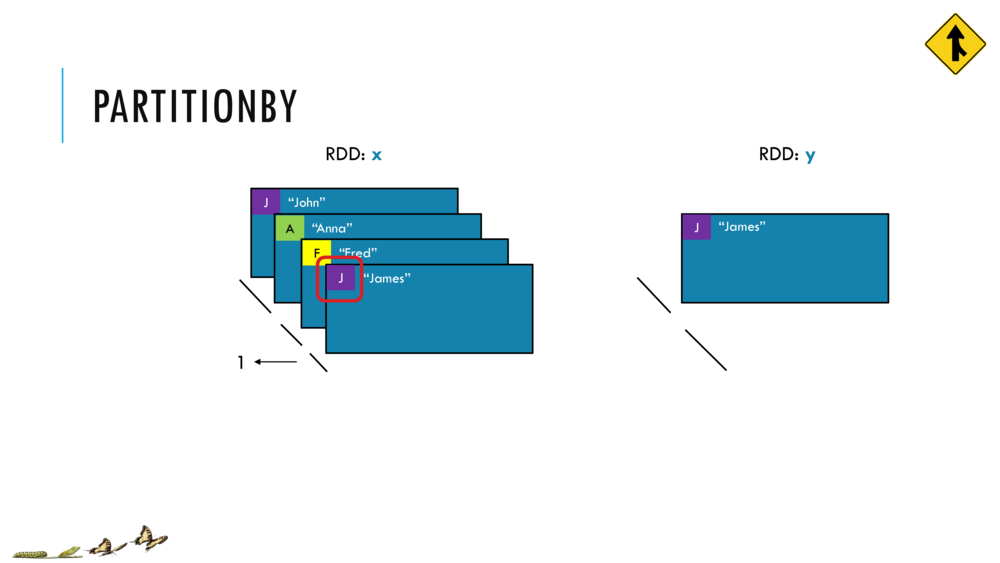
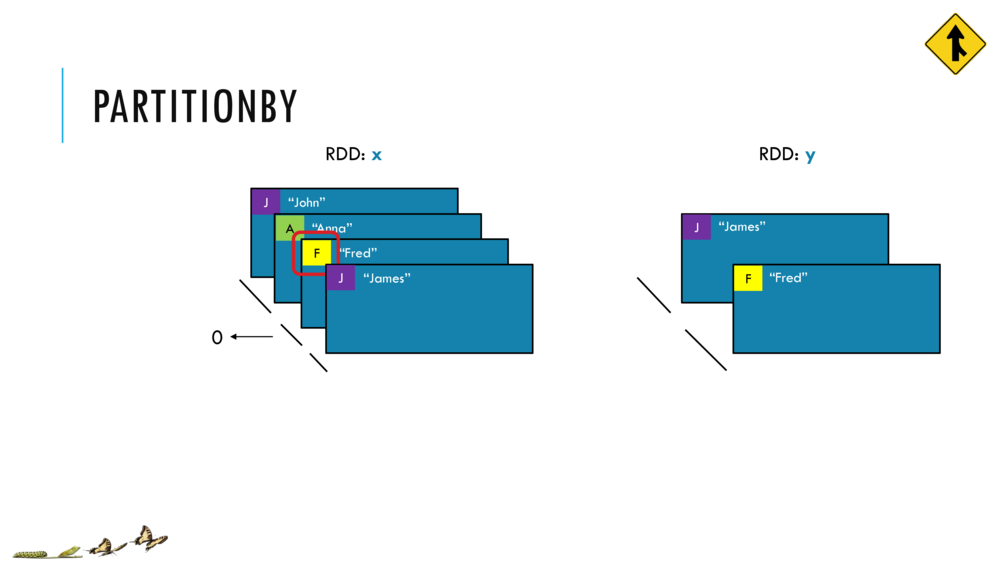
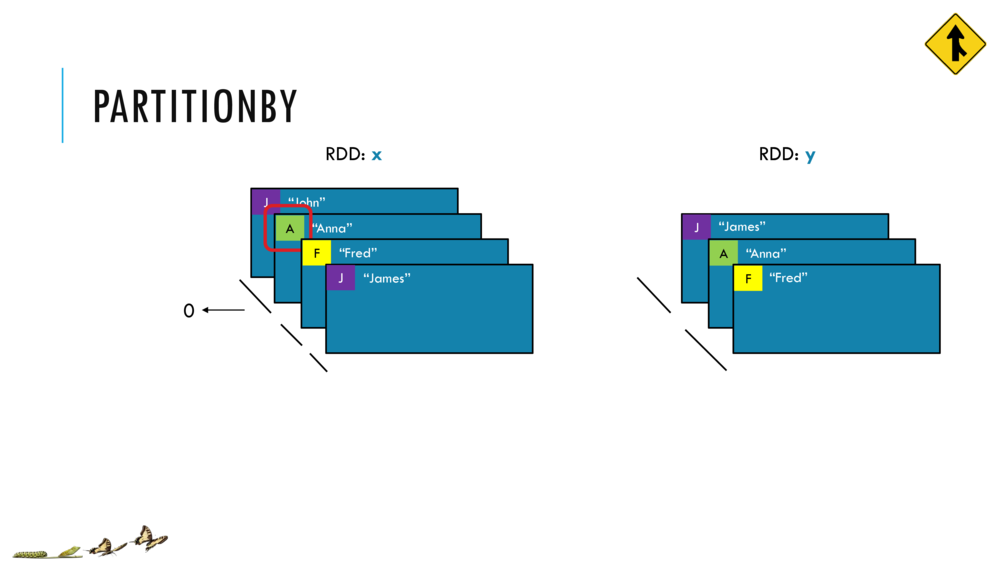
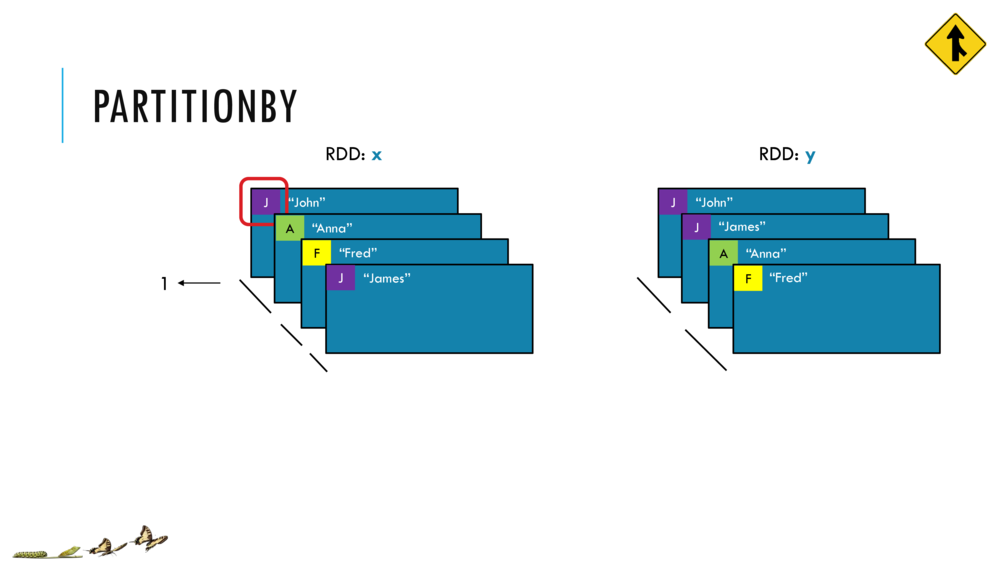
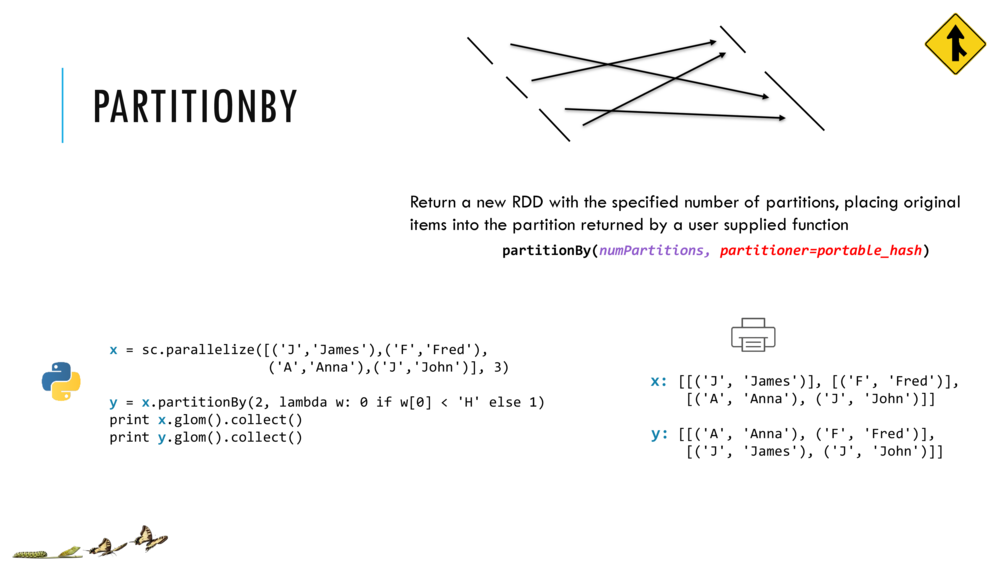
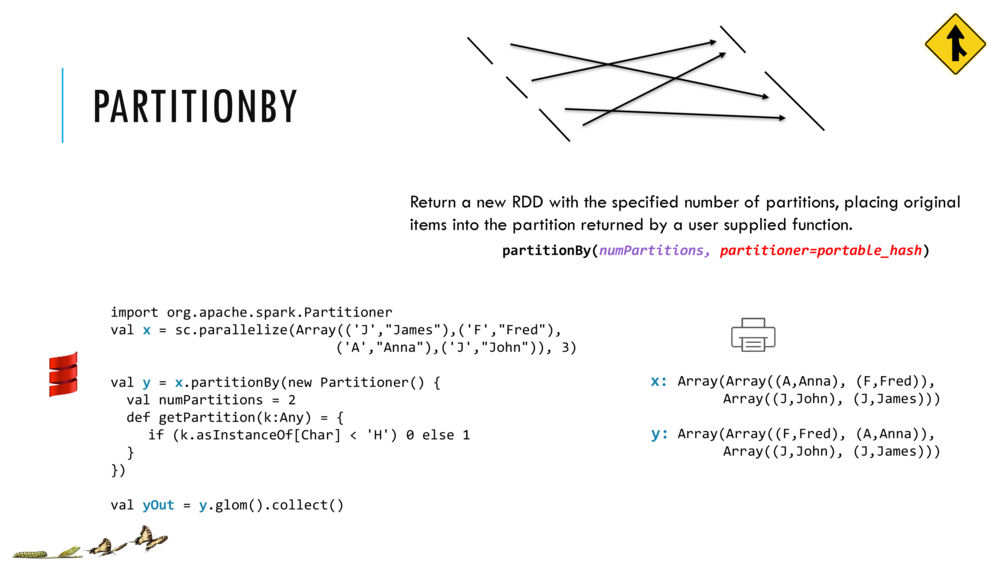
import org.apache.spark.Partitioner
import org.apache.spark.Partitioner
val x = sc.parallelize(Array(('J',"James"),('F',"Fred"), ('A',"Anna"),('J',"John")), 3)
val y = x.partitionBy(new Partitioner() {
val numPartitions = 2
def getPartition(k:Any) = {
if (k.asInstanceOf[Char] < 'H') 0 else 1
}
})
x: org.apache.spark.rdd.RDD[(Char, String)] = ParallelCollectionRDD[328] at parallelize at <console>:37
y: org.apache.spark.rdd.RDD[(Char, String)] = ShuffledRDD[329] at partitionBy at <console>:38
val yOut = y.glom().collect()
yOut: Array[Array[(Char, String)]] = Array(Array((F,Fred), (A,Anna)), Array((J,James), (J,John)))


val words = Array("one", "two", "two", "three", "three", "three")
val wordPairsRDD = sc.parallelize(words).map(word => (word, 1))
words: Array[String] = Array(one, two, two, three, three, three)
wordPairsRDD: org.apache.spark.rdd.RDD[(String, Int)] = MapPartitionsRDD[650] at map at <console>:35
val wordCountsWithReduce = wordPairsRDD
.reduceByKey(_ + _)
.collect()
wordCountsWithReduce: Array[(String, Int)] = Array((two,2), (one,1), (three,3))
val wordCountsWithGroup = wordPairsRDD
.groupByKey()
.map(t => (t._1, t._2.sum))
.collect()
wordCountsWithGroup: Array[(String, Int)] = Array((two,2), (one,1), (three,3))
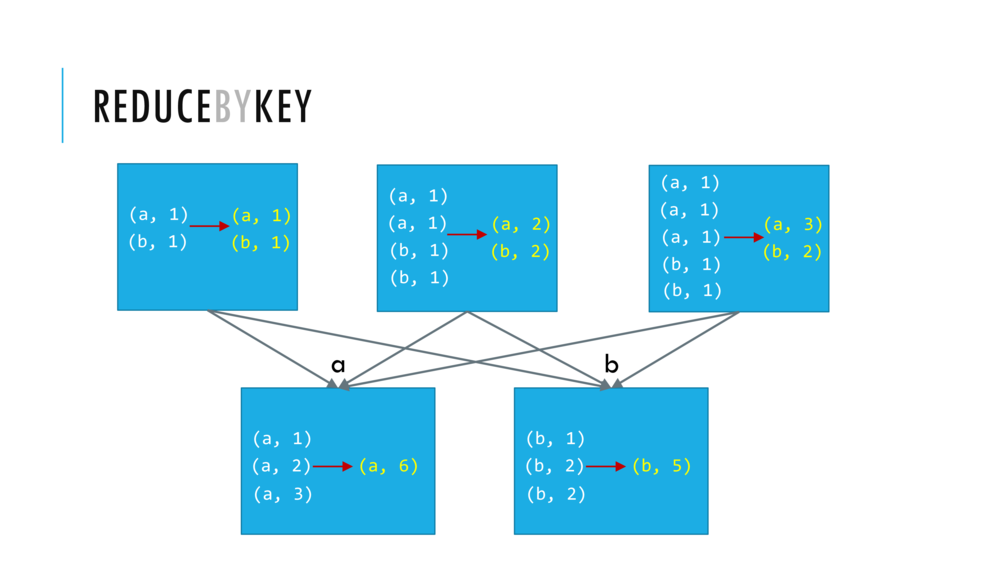
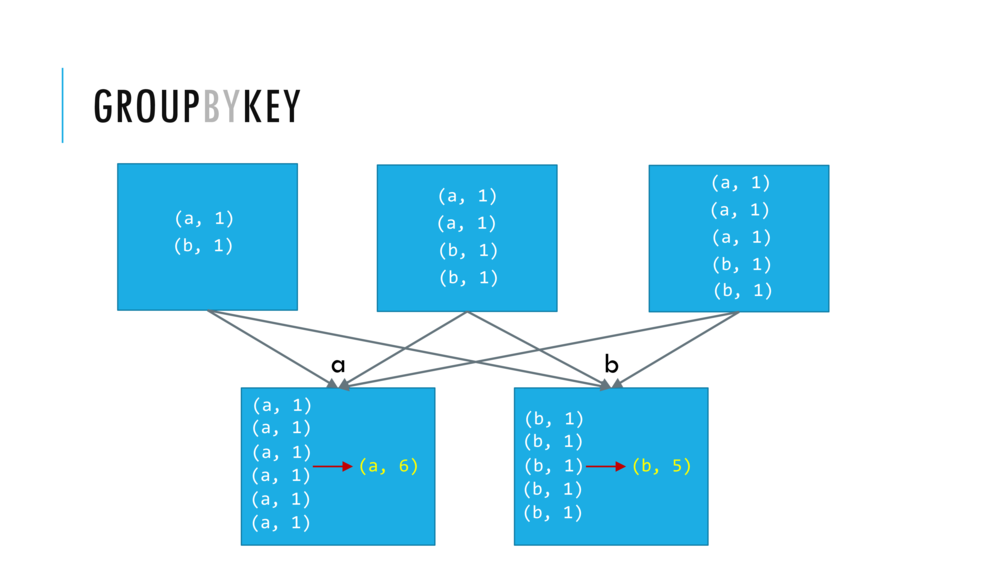
sample
Return a new RDD containing a statistical sample of the original RDD.
Let us look at the legend and overview of the visual RDD Api.
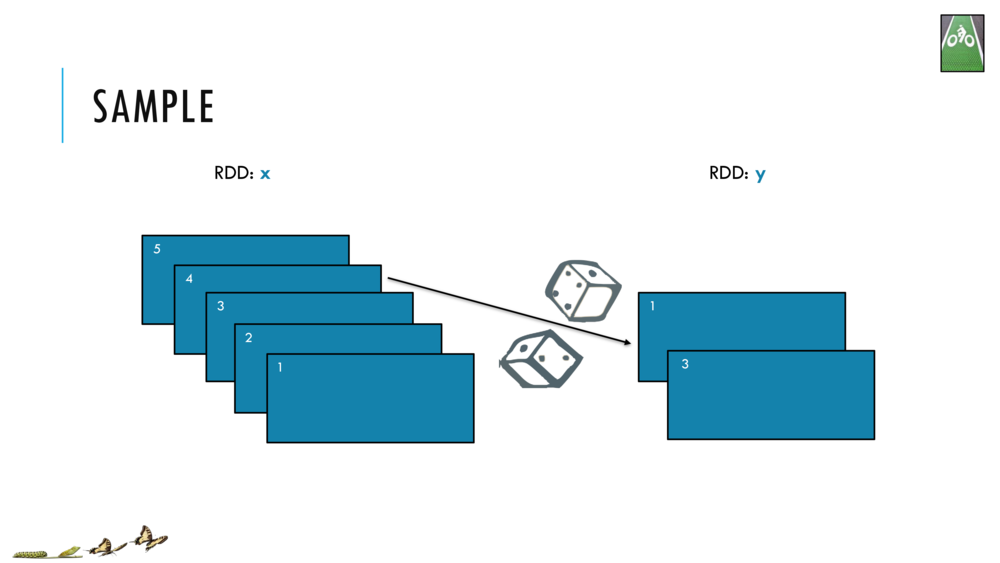
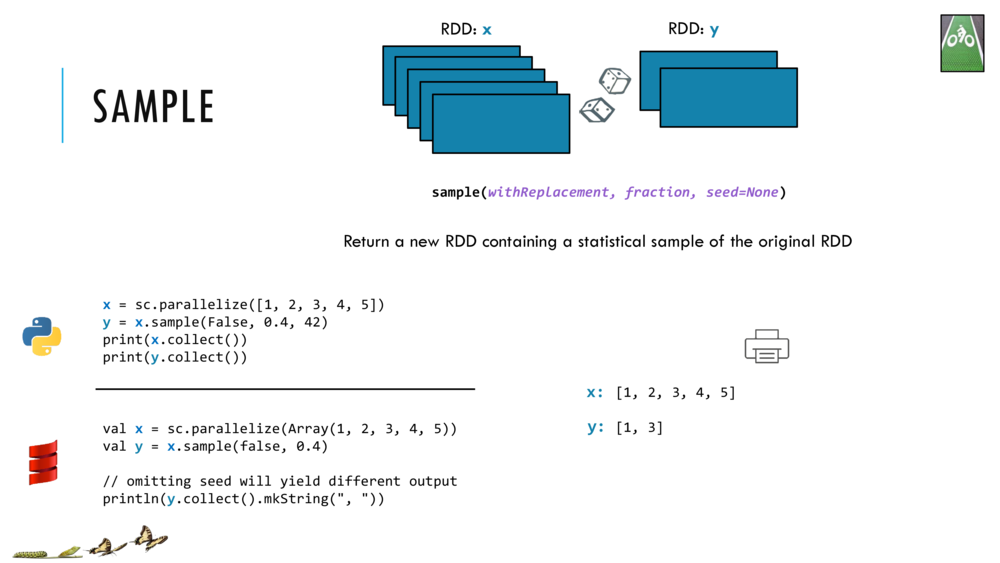
val x = sc.parallelize(Array(1, 2, 3, 4, 5))
x: org.apache.spark.rdd.RDD[Int] = ParallelCollectionRDD[789] at parallelize at <console>:34
//omitting seed will return different output
val y = x.sample(false, 0.4)
y: org.apache.spark.rdd.RDD[Int] = PartitionwiseSampledRDD[805] at sample at <console>:37
y.collect()
res1: Array[Int] = Array(2, 5)
//omitting seed will return different output
x.sample(false, 0.4).collect()
res2: Array[Int] = Array(4, 5)
//including seed will preserve output
sc.parallelize(Seq.range(0, 1000)).sample(false, 0.1, 124L).collect()
res14: Array[Int] = Array(9, 18, 27, 36, 42, 48, 49, 68, 79, 93, 94, 100, 105, 110, 114, 123, 127, 133, 135, 161, 177, 180, 181, 193, 198, 201, 222, 227, 236, 254, 263, 264, 272, 277, 287, 297, 325, 328, 336, 338, 350, 352, 353, 359, 372, 385, 409, 416, 428, 439, 443, 450, 461, 486, 489, 528, 533, 542, 543, 560, 561, 562, 563, 575, 576, 577, 578, 586, 590, 604, 614, 636, 682, 685, 723, 729, 730, 742, 749, 750, 785, 790, 809, 815, 822, 832, 865, 871, 888, 893, 906, 920, 936, 958, 982, 991, 994)
//including seed will preserve output
sc.parallelize(Seq.range(0, 1000)).sample(false, 0.1, 124L).collect()
res15: Array[Int] = Array(9, 18, 27, 36, 42, 48, 49, 68, 79, 93, 94, 100, 105, 110, 114, 123, 127, 133, 135, 161, 177, 180, 181, 193, 198, 201, 222, 227, 236, 254, 263, 264, 272, 277, 287, 297, 325, 328, 336, 338, 350, 352, 353, 359, 372, 385, 409, 416, 428, 439, 443, 450, 461, 486, 489, 528, 533, 542, 543, 560, 561, 562, 563, 575, 576, 577, 578, 586, 590, 604, 614, 636, 682, 685, 723, 729, 730, 742, 749, 750, 785, 790, 809, 815, 822, 832, 865, 871, 888, 893, 906, 920, 936, 958, 982, 991, 994)
union
Return a new RDD containing all items from two original RDDs. Duplicates are not culled (this is concatenation not set-theoretic union).
Let us look at the legend and overview of the visual RDD Api.
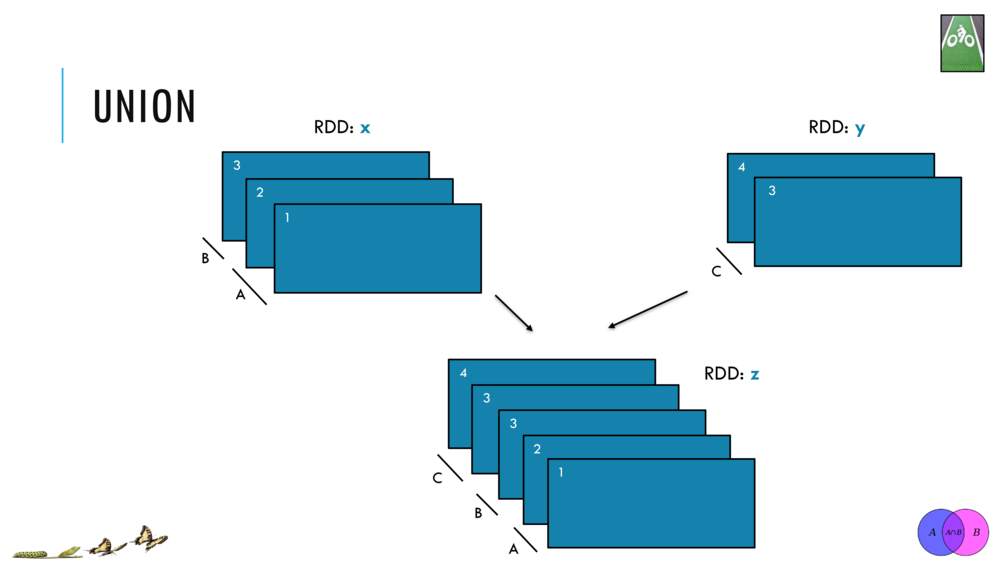
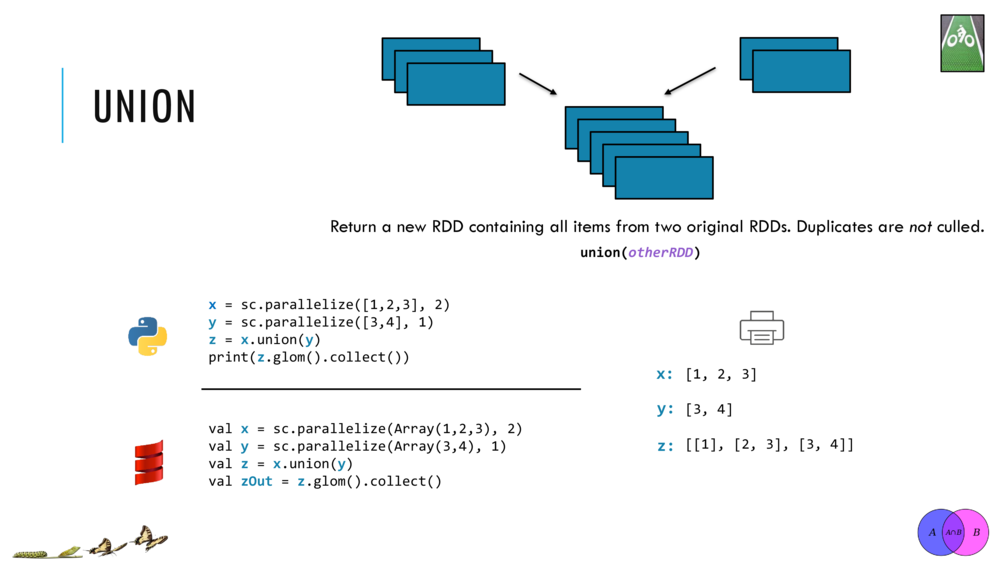
val x = sc.parallelize(Array(1, 2, 3), 2) // an RDD with 2 partitions
val y = sc.parallelize(Array(3, 4), 1) // another RDD with 1 partition
val z = x.union(y)
x: org.apache.spark.rdd.RDD[Int] = ParallelCollectionRDD[965] at parallelize at <console>:36
y: org.apache.spark.rdd.RDD[Int] = ParallelCollectionRDD[966] at parallelize at <console>:37
z: org.apache.spark.rdd.RDD[Int] = UnionRDD[967] at union at <console>:38
//glom() flattens elements on the same partition
val zOut = z.glom().collect()
zOut: Array[Array[Int]] = Array(Array(1), Array(2, 3), Array(3, 4))