// Databricks notebook source exported at Sat, 25 Jun 2016 00:18:01 UTC
Scalable Data Science
prepared by Raazesh Sainudiin and Sivanand Sivaram
The html source url of this databricks notebook and its recorded Uji  :
:
#Power Plant ML Pipeline Application This is an end-to-end example of using a number of different machine learning algorithms to solve a supervised regression problem.
###Table of Contents
- Step 1: Business Understanding
- Step 2: Load Your Data
- Step 3: Explore Your Data
- Step 4: Visualize Your Data
- Step 5: Data Preparation
- Step 6: Data Modeling
- Step 7: Tuning and Evaluation
- Step 8: Deployment
We are trying to predict power output given a set of readings from various sensors in a gas-fired power generation plant. Power generation is a complex process, and understanding and predicting power output is an important element in managing a plant and its connection to the power grid.
More information about Peaker or Peaking Power Plants can be found on Wikipedia https://en.wikipedia.org/wiki/Peaking_power_plant
Given this business problem, we need to translate it to a Machine Learning task. The ML task is regression since the label (or target) we are trying to predict is numeric.
The example data is provided by UCI at UCI Machine Learning Repository Combined Cycle Power Plant Data Set
You can read the background on the UCI page, but in summary we have collected a number of readings from sensors at a Gas Fired Power Plant
(also called a Peaker Plant) and now we want to use those sensor readings to predict how much power the plant will generate.
More information about Machine Learning with Spark can be found in the Spark MLLib Programming Guide
Please note this example only works with Spark version 1.4 or higher
To Rerun Steps 1-4 done in the notebook at:
You have the following two options:
- Rerun from here:
- Copy the following
%run "..."command into the cell below and evaluate it by pressing Ctrl+Enter
- Copy the following
%run "/scalable-data-science/week3/05_SparkSQLETLEDA/009_PowerPlantPipeline_01ETLEDA"
- Note: If you already evaluated the
%run ...command above then:- first delete the cell by pressing on
xon the top-right corner of the cell and - revaluate it as instructed in Rerun from here
- first delete the cell by pressing on
- Rerun from there: simply go to this notebook by right-clicking and opening in New Tab:
- Workspace -> scalable-data-science -> week3 -> 05_SparkSQLETLEDA -> 009_PowerPlantPipeline_01ETLEDA
- and click
Run Allat the top of that notebook.
- Note: We have already evaluated the
%run ...command below. Therefore, you need to:- first delete the cell below by pressing on
xon the top-right corner of the cell and - revaluate it as instructed in Rerun from here instruction above.
- first delete the cell below by pressing on
%run "/scalable-data-science/week3/05_SparkSQLETLEDA/009_PowerPlantPipeline_01ETLEDA" // use right path!
Step 5: Data Preparation, Step 6: Modeling, Step 7: Tuning and Evaluation, and Step 8: Deployment
##Step 5: Data Preparation
The next step is to prepare the data. Since all of this data is numeric and consistent, this is a simple task for us today.
We will need to convert the predictor features from columns to Feature Vectors using the org.apache.spark.ml.feature.VectorAssembler
The VectorAssembler will be the first step in building our ML pipeline.
//Let's quickly recall the schema
table("power_plant_table").printSchema
NOTE: powerPlantDF declared in notebook 009_PowerPlantPipeline_01ETLEDA will not be availble in this notebook if you chose option 2 earlier and clicked ‘Run All’ directly after opening the notebook 009_PowerPlantPipeline_01ETLEDA instead of option 1 via:
%run "/scalable-data-science/week3/05_SparkSQLETLEDA/009_PowerPlantPipeline_01ETLEDA"
This because DataFrames are notebook-specific but tables are shard-specific.
// powerPlantDF
import org.apache.spark.ml.feature.VectorAssembler
// make a DataFrame called dataset from the table
val dataset = sqlContext.table("power_plant_table")
val vectorizer = new VectorAssembler()
.setInputCols(Array("AT", "V", "AP", "RH"))
.setOutputCol("features")
##Step 6: Data Modeling Now let’s model our data to predict what the power output will be given a set of sensor readings
Our first model will be based on simple linear regression since we saw some linear patterns in our data based on the scatter plots during the exploration stage.
%run "/scalable-data-science/xtraResources/support/sdsFunctions"
Linear Regression Model
- Linear Regression is one of the most useful work-horses of statistical learning
- See Chapter 7 of Kevin Murphy’s Machine Learning froma Probabilistic Perspective for a good mathematical and algorithmic introduction.
- You should have already seen Ameet’s treatment of the topic from earlier notebook.
Let’s open http://spark.apache.org/docs/latest/mllib-linear-methods.html#regression for some details.
//This allows easy embedding of publicly available information into any other notebook
//when viewing in git-book just ignore this block - you may have to manually chase the URL in frameIt("URL").
//Example usage:
// displayHTML(frameIt("https://en.wikipedia.org/wiki/Latent_Dirichlet_allocation#Topics_in_LDA",250))
def frameIt( u:String, h:Int ) : String = {
"""<iframe
src=""""+ u+""""
width="95%" height="""" + h + """"
sandbox>
<p>
<a href="http://spark.apache.org/docs/latest/index.html">
Fallback link for browsers that, unlikely, don't support frames
</a>
</p>
</iframe>"""
}
displayHTML(frameIt("http://spark.apache.org/docs/latest/mllib-linear-methods.html#regression",300))
// First let's hold out 20% of our data for testing and leave 80% for training
var Array(split20, split80) = dataset.randomSplit(Array(0.20, 0.80), 1800009193L)
A Note on being able to link-in scala docs from URL
Unfortunately we cannot deep-link-in to scala docs as explained here: http://www.scala-lang.org/old/node/8798.html.
Our best option now seems to go to the following link and search for randomSplit in the search-box there:
The syntax for randomSplit (obtained by converting the source of the highlighted html resulting from the search from the link above and converting it to markdown via pandoc in bash) is as follows:
def randomSplit(weights: Array[Double], seed: Long): Array[DataFrame]
Randomly splits this DataFrame with the provided weights.
- weights
- weights for splits, will be normalized if they don’t sum to 1.
- seed
- Seed for sampling.
- Since
- 1.4.0
Python is ok http://spark.apache.org/docs/latest/api/python/pyspark.sql.html#pyspark.sql.DataFrame.randomSplit
Java too http://spark.apache.org/docs/latest/api/java/org/apache/spark/sql/DataFrame.html#randomSplit(double%5B%5D%2C%20long)
// Let's cache these datasets for performance
val testSet = split20.cache()
val trainingSet = split80.cache()
dataset.take(3)
testSet.take(3)
trainingSet.take(3)
// ***** LINEAR REGRESSION MODEL ****
import org.apache.spark.ml.regression.LinearRegression
import org.apache.spark.ml.regression.LinearRegressionModel
import org.apache.spark.ml.Pipeline
// Let's initialize our linear regression learner
val lr = new LinearRegression()
// We use explain params to dump the parameters we can use
lr.explainParams()
The cell below is based on the Spark ML pipeline API. More information can be found in the Spark ML Programming Guide at https://spark.apache.org/docs/latest/ml-guide.html
// Now we set the parameters for the method
lr.setPredictionCol("Predicted_PE")
.setLabelCol("PE")
.setMaxIter(100)
.setRegParam(0.1)
// We will use the new spark.ml pipeline API. If you have worked with scikit-learn this will be very familiar.
val lrPipeline = new Pipeline()
lrPipeline.setStages(Array(vectorizer, lr))
// Let's first train on the entire dataset to see what we get
val lrModel = lrPipeline.fit(trainingSet)
Since Linear Regression is simply a line of best fit over the data that minimizes the square of the error, given multiple input dimensions we can express each predictor as a line function of the form:
\[y = b_0 + b_1 x_1 + b_2 x_2 + b_3 x_3 + \ldots + b_i x_i + \ldots + b_k x_k\]where \(b_0\) is the intercept and \(b_i\)’s are coefficients.
To express the coefficients of that line we can retrieve the Estimator stage from the fitted, linear-regression pipeline model named lrModel and express the weights and the intercept for the function.
// The intercept is as follows:
val intercept = lrModel.stages(1).asInstanceOf[LinearRegressionModel].intercept
// The coefficents (i.e. weights) are as follows:
val weights = lrModel.stages(1).asInstanceOf[LinearRegressionModel].coefficients.toArray
The model has been fit and the intercept and coefficients displayed above.
Now, let us do some work to make a string of the model that is easy to understand for an applied data scientist or data analyst.
val featuresNoLabel = dataset.columns.filter(col => col != "PE")
val coefficentFeaturePairs = sc.parallelize(weights).zip(sc.parallelize(featuresNoLabel))
coefficentFeaturePairs.collect() // this just pairs each coefficient with the name of its corresponding feature
// Now let's sort the coefficients from the largest to the smallest
var equation = s"y = $intercept "
//var variables = Array
coefficentFeaturePairs.sortByKey().collect().foreach({
case (weight, feature) =>
{
val symbol = if (weight > 0) "+" else "-"
val absWeight = Math.abs(weight)
equation += (s" $symbol (${absWeight} * ${feature})")
}
}
)
// Finally here is our equation
println("Linear Regression Equation: " + equation)
Based on examining the fitted Linear Regression Equation above:
- There is a strong negative correlation between Atmospheric Temperature (AT) and Power Output due to the coefficient being greater than -1.91.
- But our other dimenensions seem to have little to no correlation with Power Output.
Do you remember Step 2: Explore Your Data? When we visualized each predictor against Power Output using a Scatter Plot, only the temperature variable seemed to have a linear correlation with Power Output so our final equation seems logical.
Now let’s see what our predictions look like given this model.
val predictionsAndLabels = lrModel.transform(testSet)
display(predictionsAndLabels.select("AT", "V", "AP", "RH", "PE", "Predicted_PE"))
Now that we have real predictions we can use an evaluation metric such as Root Mean Squared Error to validate our regression model. The lower the Root Mean Squared Error, the better our model.
//Now let's compute some evaluation metrics against our test dataset
import org.apache.spark.mllib.evaluation.RegressionMetrics
val metrics = new RegressionMetrics(predictionsAndLabels.select("Predicted_PE", "PE").rdd.map(r => (r(0).asInstanceOf[Double], r(1).asInstanceOf[Double])))
val rmse = metrics.rootMeanSquaredError
val explainedVariance = metrics.explainedVariance
val r2 = metrics.r2
println (f"Root Mean Squared Error: $rmse")
println (f"Explained Variance: $explainedVariance")
println (f"R2: $r2")
Generally a good model will have 68% of predictions within 1 RMSE and 95% within 2 RMSE of the actual value. Let’s calculate and see if our RMSE meets this criteria.
display(predictionsAndLabels) // recall the DataFrame predictionsAndLabels
// First we calculate the residual error and divide it by the RMSE from predictionsAndLabels DataFrame and make another DataFrame that is registered as a temporary table Power_Plant_RMSE_Evaluation
predictionsAndLabels.selectExpr("PE", "Predicted_PE", "PE - Predicted_PE AS Residual_Error", s""" (PE - Predicted_PE) / $rmse AS Within_RSME""").registerTempTable("Power_Plant_RMSE_Evaluation")
%sql SELECT * from Power_Plant_RMSE_Evaluation
%sql -- Now we can display the RMSE as a Histogram. Clearly this shows that the RMSE is centered around 0 with the vast majority of the error within 2 RMSEs.
SELECT Within_RSME from Power_Plant_RMSE_Evaluation
We can see this definitively if we count the number of predictions within + or - 1.0 and + or - 2.0 and display this as a pie chart:
%sql
SELECT case when Within_RSME <= 1.0 and Within_RSME >= -1.0 then 1 when Within_RSME <= 2.0 and Within_RSME >= -2.0 then 2 else 3 end RSME_Multiple, COUNT(*) count from Power_Plant_RMSE_Evaluation
group by case when Within_RSME <= 1.0 and Within_RSME >= -1.0 then 1 when Within_RSME <= 2.0 and Within_RSME >= -2.0 then 2 else 3 end
So we have about 70% of our training data within 1 RMSE and about 98% (70% + 29%) within 2 RMSE. So the model is pretty decent. Let’s see if we can tune the model to improve it further.
NOTE: these numbers will vary across runs due to the seed in random sampling of training and test set, number of iterations, and other stopping rules in optimization, for example.
#Step 7: Tuning and Evaluation
Now that we have a model with all of the data let’s try to make a better model by tuning over several parameters.
import org.apache.spark.ml.tuning.{ParamGridBuilder, CrossValidator}
import org.apache.spark.ml.evaluation._
displayHTML(frameIt("http://spark.apache.org/docs/latest/ml-guide.html#example-model-selection-via-cross-validation",500))
First let’s use a cross validator to split the data into training and validation subsets. See ml-guide.html#example-model-selection-via-cross-validation.
//Let's set up our evaluator class to judge the model based on the best root mean squared error
val regEval = new RegressionEvaluator()
regEval.setLabelCol("PE")
.setPredictionCol("Predicted_PE")
.setMetricName("rmse")
We now treat the lrPipeline as an Estimator, wrapping it in a CrossValidator instance.
This will allow us to jointly choose parameters for all Pipeline stages.
A CrossValidator requires an Estimator, an Evaluator (which we set next).
//Let's create our crossvalidator with 3 fold cross validation
val crossval = new CrossValidator()
crossval.setEstimator(lrPipeline)
crossval.setNumFolds(3)
crossval.setEvaluator(regEval)
A CrossValidator also requires a set of EstimatorParamMaps which we set next.
For this we need a regularization parameter (more generally a hyper-parameter that is model-specific).
Now, let’s tune over our regularization parameter from 0.01 to 0.10.
val regParam = ((1 to 10) toArray).map(x => (x /100.0))
Check out the scala docs for syntactic details on org.apache.spark.ml.tuning.ParamGridBuilder.
val paramGrid = new ParamGridBuilder()
.addGrid(lr.regParam, regParam)
.build()
crossval.setEstimatorParamMaps(paramGrid)
//Now let's create our model
val cvModel = crossval.fit(trainingSet)
In addition to CrossValidator Spark also offers TrainValidationSplit for hyper-parameter tuning. TrainValidationSplit only evaluates each combination of parameters once as opposed to k times in case of CrossValidator. It is therefore less expensive, but will not produce as reliable results when the training dataset is not sufficiently large.
- http://spark.apache.org/docs/latest/ml-guide.html#example-model-selection-via-train-validation-split
Now that we have tuned let’s see what we got for tuning parameters and what our RMSE was versus our intial model
val predictionsAndLabels = cvModel.transform(testSet)
val metrics = new RegressionMetrics(predictionsAndLabels.select("Predicted_PE", "PE").rdd.map(r => (r(0).asInstanceOf[Double], r(1).asInstanceOf[Double])))
val rmse = metrics.rootMeanSquaredError
val explainedVariance = metrics.explainedVariance
val r2 = metrics.r2
println (f"Root Mean Squared Error: $rmse")
println (f"Explained Variance: $explainedVariance")
println (f"R2: $r2")
So our initial untuned and tuned linear regression models are statistically identical.
Given that the only linearly correlated variable is Temperature, it makes sense try another machine learning method such a Decision Tree to handle non-linear data and see if we can improve our model
A Decision Tree creates a model based on splitting variables using a tree structure. We will first start with a single decision tree model.
Reference Decision Trees: https://en.wikipedia.org/wiki/Decision_tree_learning
//Let's build a decision tree pipeline
import org.apache.spark.ml.regression.DecisionTreeRegressor
val dt = new DecisionTreeRegressor()
dt.setLabelCol("PE")
dt.setPredictionCol("Predicted_PE")
dt.setFeaturesCol("features")
dt.setMaxBins(100)
val dtPipeline = new Pipeline()
dtPipeline.setStages(Array(vectorizer, dt))
//Let's just resuse our CrossValidator
crossval.setEstimator(dtPipeline)
val paramGrid = new ParamGridBuilder()
.addGrid(dt.maxDepth, Array(2, 3))
.build()
crossval.setEstimatorParamMaps(paramGrid)
val dtModel = crossval.fit(trainingSet) // fit decitionTree with cv
import org.apache.spark.ml.regression.DecisionTreeRegressionModel
import org.apache.spark.ml.PipelineModel
dtModel.bestModel.asInstanceOf[PipelineModel].stages.last.asInstanceOf[DecisionTreeRegressionModel].toDebugString
The line above will pull the Decision Tree model from the Pipeline and display it as an if-then-else string.
Next let’s visualize it as a decision tree for regression.
display(dtModel.bestModel.asInstanceOf[PipelineModel].stages.last.asInstanceOf[DecisionTreeRegressionModel])
Now let’s see how our DecisionTree model compares to our LinearRegression model
val predictionsAndLabels = dtModel.bestModel.transform(testSet)
val metrics = new RegressionMetrics(predictionsAndLabels.select("Predicted_PE", "PE").map(r => (r(0).asInstanceOf[Double], r(1).asInstanceOf[Double])))
val rmse = metrics.rootMeanSquaredError
val explainedVariance = metrics.explainedVariance
val r2 = metrics.r2
println (f"Root Mean Squared Error: $rmse")
println (f"Explained Variance: $explainedVariance")
println (f"R2: $r2")
So our DecisionTree was slightly worse than our LinearRegression model (LR: 4.51 vs DT: 5.03). Maybe we can try an Ensemble method such as Gradient-Boosted Decision Trees to see if we can strengthen our model by using an ensemble of weaker trees with weighting to reduce the error in our model.
Note since this is a complex model, the cell below can take about *16 minutes or so to run, go out and grab a coffee and come back :-).*
DON’T run the next 3 cells in class!
The KDnuggets site: http://www.kdnuggets.com/2015/05/top-10-data-mining-algorithms-explained.html is recommended by Andrey Konstantinov, a ChristchurchSparker, and is good for a quick high-level insights about varion ML algorithms.
Let’s see what a boosting algorithm, a type of ensemble method, is all about from here.
displayHTML(frameIt("http://www.kdnuggets.com/2015/05/top-10-data-mining-algorithms-explained.html/3",700))
displayHTML(frameIt("http://spark.apache.org/docs/latest/mllib-ensembles.html#gradient-boosted-trees-gbts",500))
This can take over 400 to 500 seconds!
import org.apache.spark.ml.regression.GBTRegressor
val gbt = new GBTRegressor()
gbt.setLabelCol("PE")
gbt.setPredictionCol("Predicted_PE")
gbt.setFeaturesCol("features")
gbt.setSeed(100088121L)
gbt.setMaxBins(100)
gbt.setMaxIter(120)
val gbtPipeline = new Pipeline()
gbtPipeline.setStages(Array(vectorizer, gbt))
//Let's just resuse our CrossValidator
crossval.setEstimator(gbtPipeline)
val paramGrid = new ParamGridBuilder()
.addGrid(gbt.maxDepth, Array(2, 3))
.build()
crossval.setEstimatorParamMaps(paramGrid)
//gbt.explainParams
val gbtModel = crossval.fit(trainingSet)
import org.apache.spark.ml.regression.GBTRegressionModel
val predictionsAndLabels = gbtModel.bestModel.transform(testSet)
val metrics = new RegressionMetrics(predictionsAndLabels.select("Predicted_PE", "PE").map(r => (r(0).asInstanceOf[Double], r(1).asInstanceOf[Double])))
val rmse = metrics.rootMeanSquaredError
val explainedVariance = metrics.explainedVariance
val r2 = metrics.r2
println (f"Root Mean Squared Error: $rmse")
println (f"Explained Variance: $explainedVariance")
println (f"R2: $r2")
Note that the root mean squared error is much smaller now due to the ensemble of 120 trees from Gradient Boosting!
We can use the toDebugString method to dump out what our trees and weighting look like:
gbtModel.bestModel.asInstanceOf[PipelineModel].stages.last.asInstanceOf[GBTRegressionModel].toDebugString
### Conclusion
Wow! So our best model is in fact our Gradient Boosted Decision tree model which uses an ensemble of 120 Trees with a depth of 3 to construct a better model than the single decision tree.
#Step 8: Deployment
Now that we have a predictive model it is time to deploy the model into an operational environment.
In our example, let’s say we have a series of sensors attached to the power plant and a monitoring station.
The monitoring station will need close to real-time information about how much power that their station will generate so they can relay that to the utility.
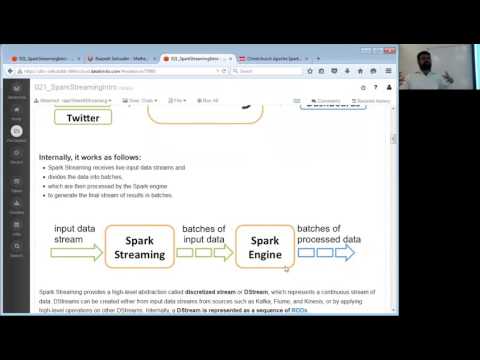
So let’s create a Spark Streaming utility that we can use for this purpose.
See http://spark.apache.org/docs/latest/streaming-programming-guide.html if you can’t wait!
After deployment you will be able to use the best predictions from gradient boosed regression trees to feed a real-time dashboard or feed the utility with information on how much power the peaker plant will deliver give current conditions.
// Let's set the variable finalModel to our best GBT Model
val finalModel = gbtModel.bestModel
Let’s create our table for predictions
%sql
DROP TABLE IF EXISTS power_plant_predictions ;
CREATE TABLE power_plant_predictions(
AT Double,
V Double,
AP Double,
RH Double,
PE Double,
Predicted_PE Double
);
Now let’s create our streaming job to score new power plant readings in real-time.
CAUTION: There can be only one spark streaming context per cluster!!! So please check if a streaming context is already alive first.
import java.nio.ByteBuffer
import java.net._
import java.io._
import concurrent._
import scala.io._
import sys.process._
import org.apache.spark.Logging
import org.apache.spark.SparkConf
import org.apache.spark.storage.StorageLevel
import org.apache.spark.streaming.Seconds
import org.apache.spark.streaming.Minutes
import org.apache.spark.streaming.StreamingContext
import org.apache.spark.streaming.StreamingContext.toPairDStreamFunctions
import org.apache.log4j.Logger
import org.apache.log4j.Level
import org.apache.spark.streaming.receiver.Receiver
import sqlContext._
import net.liftweb.json.DefaultFormats
import net.liftweb.json._
import scala.collection.mutable.SynchronizedQueue
val queue = new SynchronizedQueue[RDD[String]]()
val batchIntervalSeconds = 2
var newContextCreated = false // Flag to detect whether new context was created or not
// Function to create a new StreamingContext and set it up
def creatingFunc(): StreamingContext = {
// Create a StreamingContext
val ssc = new StreamingContext(sc, Seconds(batchIntervalSeconds))
val batchInterval = Seconds(1)
ssc.remember(Seconds(300))
val dstream = ssc.queueStream(queue)
dstream.foreachRDD {
rdd =>
// if the RDD has data
if(!(rdd.isEmpty())) {
// Use the final model to transform a JSON message into a dataframe and pass the dataframe to our model's transform method
finalModel
.transform(read.json(rdd).toDF())
// Select only columns we are interested in
.select("AT", "V", "AP", "RH", "PE", "Predicted_PE")
// Append the results to our power_plant_predictions table
.write.mode(SaveMode.Append).saveAsTable("power_plant_predictions")
}
}
println("Creating function called to create new StreamingContext for Power Plant Predictions")
newContextCreated = true
ssc
}
val ssc = StreamingContext.getActiveOrCreate(creatingFunc)
if (newContextCreated) {
println("New context created from currently defined creating function")
} else {
println("Existing context running or recovered from checkpoint, may not be running currently defined creating function")
}
ssc.start()
Now that we have created and defined our streaming job, let’s test it with some data. First we clear the predictions table.
%sql truncate table power_plant_predictions
Let’s use data to see how much power output our model will predict.
// First we try it with a record from our test set and see what we get:
queue += sc.makeRDD(Seq(s"""{"AT":10.82,"V":37.5,"AP":1009.23,"RH":96.62,"PE":473.9}"""))
// We may need to wait a few seconds for data to appear in the table
Thread.sleep(Seconds(5).milliseconds)
%sql
--and we can query our predictions table
select * from power_plant_predictions
Let’s repeat with a different test measurement that our model has not seen before:
queue += sc.makeRDD(Seq(s"""{"AT":10.0,"V":40,"AP":1000,"RH":90.0,"PE":0.0}"""))
Thread.sleep(Seconds(5).milliseconds)
%sql
--Note you may have to run this a couple of times to see the refreshed data...
select * from power_plant_predictions
As you can see the Predictions are very close to the real data points.
%sql
select * from power_plant_table where AT between 10 and 11 and AP between 1000 and 1010 and RH between 90 and 97 and v between 37 and 40 order by PE
Now you use the predictions table to feed a real-time dashboard or feed the utility with information on how much power the peaker plant will deliver.
Make sure the streaming context is stopped when you are done, as there can be only one such context per cluster!
ssc.stop(stopSparkContext = false) // gotto stop or it ill keep running!!!
Datasource References:
- Pinar Tüfekci, Prediction of full load electrical power output of a base load operated combined cycle power plant using machine learning methods, International Journal of Electrical Power & Energy Systems, Volume 60, September 2014, Pages 126-140, ISSN 0142-0615, Web Link
- Heysem Kaya, Pinar Tüfekci , Sadik Fikret Gürgen: Local and Global Learning Methods for Predicting Power of a Combined Gas & Steam Turbine, Proceedings of the International Conference on Emerging Trends in Computer and Electronics Engineering ICETCEE 2012, pp. 13-18 (Mar. 2012, Dubai) Web Link